

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2022, Vol. 42 ›› Issue (6): 705-715.DOI: 10.16829/j.slxb.150645
赵琪1, 张琪1, 李浩玲1, 兰月2, 鄢行安3, 赵贵军3( ), 戚文华1(
), 戚文华1( )
)
收稿日期:2021-12-01
接受日期:2022-07-05
出版日期:2022-11-30
发布日期:2022-12-02
通讯作者:
赵贵军,戚文华
作者简介:赵琪 (1996- ),女,硕士研究生,主要从事动植物分子/遗传资源利用研究;基金资助:
Qi ZHAO1, Qi ZHANG1, Haoling LI1, Yue LAN2, Xingan YAN3, Guijun ZHAO3( ), Wenhua QI1(
), Wenhua QI1( )
)
Received:2021-12-01
Accepted:2022-07-05
Online:2022-11-30
Published:2022-12-02
Contact:
Guijun ZHAO,Wenhua QI
摘要:
麝科和鹿科动物均属于偶蹄反刍类动物,具有重要的经济价值。通过系统的微卫星序列 (Simple sequences repeats, SSRs) 从基因组水平揭示物种间的系统进化关系,探索微卫星序列的基因功能及其富集的信号通路,目前仍缺乏相关研究。随着林麝 (Moschus berezovskii)、原麝 (Moschus moschiferus)、小麂 (Muntiacus reevesi)、赤麂 (Muntiacus vaginalis) 和马鹿 (Cervus elaphus) 基因组测序的完成,本文利用生物信息学方法提取了这些动物蛋白质编码区 (coding sequences, CDS) 序列,统计和分析了其CDS区微卫星序列分布规律及其生物学功能,探索了含SSR 基因富集的信号通路及其与疾病的关联性。结果表明,林麝、原麝、小麂、赤麂和马鹿蛋白质编码区含SSR序列的基因所占比例分别为6.96% (1 696个)、7.18% (2 359个)、7.29% (3 005个)、7.36% (1 916个) 和7.48% (1 924个),并且这5种动物CDS区SSRs分布模式具有相似性,均是三倍体核苷酸 (即三核苷酸和六核苷酸) SSRs最多,分别为96.85%、94.87%、65.44%、64.23%和88.04%。GO功能富集表明,林麝与其他4种动物蛋白质编码区SSR序列在分子功能、细胞组成和生物学过程3个方面具有较多共同显著富集的功能,包括DNA结合、染色质和生长发育等。KEGG 通路分析表明,林麝及其他4种动物蛋白质编码区SSR序列具有7个共同显著富集的KEGG通路,包括遗传信息调控蛋白家族、转录因子、染色体及相关蛋白、剪接体、转录机制和Notch信号通路和成体糖尿病。通过对林麝编码区含SSR关键免疫基因及其相关联的KEGG通路进行分析,发现10个含SSR的关键免疫基因对应的KEGG通路与疾病密切相关。
中图分类号:
赵琪, 张琪, 李浩玲, 兰月, 鄢行安, 赵贵军, 戚文华. 林麝及其近缘物种编码区微卫星分布规律及功能分析[J]. 兽类学报, 2022, 42(6): 705-715.
Qi ZHAO, Qi ZHANG, Haoling LI, Yue LAN, Xingan YAN, Guijun ZHAO, Wenhua QI. Distinct patterns of microsatellite and functional analysis of forest musk deer and its closely related species[J]. ACTA THERIOLOGICA SINICA, 2022, 42(6): 705-715.
| 参数 Parameters | 林麝 Moschus berezovskii | 原麝 Moschus moschiferus | 小麂 Muntiacusreeves | 赤麂 Muntiacus vaginalis | 马鹿 Cervus elaphus |
|---|---|---|---|---|---|
| 编码区基因数量 Total number of genes of CDS | 24 352 | 29 305 | 26 044 | 25 737 | 28 103 |
| GC含量 GC content (%) | 53.79 | 53.37 | 53.34 | 53.41 | 53.60 |
| SSR序列数量 Number of SSRs | 2 542 | 3 118 | 2 769 | 2 726 | 3 729 |
| SSR序列长度 Total length of SSRs (bp) | 49 233 | 76 969 | 85 507 | 708 052 | 67 743 |
| SSR序列占编码区序列的比例 CDS SSRs content (%) | 6.96 | 7.18 | 7.36 | 7.48 | 7.29 |
| 丰度 Relative abundance (No./Mb) | 75.28 | 64.63 | 65.18 | 60.86 | 52.64 |
| 密度 Relative density (bp/Mb) | 1 432.07 | 1 194.42 | 1 169.14 | 1 084.19 | 10 248.16 |
表1 林麝及其近缘物种编码区基因概况
Table 1 Overview of the CDS of Moschus berezovskii and its closely related species
| 参数 Parameters | 林麝 Moschus berezovskii | 原麝 Moschus moschiferus | 小麂 Muntiacusreeves | 赤麂 Muntiacus vaginalis | 马鹿 Cervus elaphus |
|---|---|---|---|---|---|
| 编码区基因数量 Total number of genes of CDS | 24 352 | 29 305 | 26 044 | 25 737 | 28 103 |
| GC含量 GC content (%) | 53.79 | 53.37 | 53.34 | 53.41 | 53.60 |
| SSR序列数量 Number of SSRs | 2 542 | 3 118 | 2 769 | 2 726 | 3 729 |
| SSR序列长度 Total length of SSRs (bp) | 49 233 | 76 969 | 85 507 | 708 052 | 67 743 |
| SSR序列占编码区序列的比例 CDS SSRs content (%) | 6.96 | 7.18 | 7.36 | 7.48 | 7.29 |
| 丰度 Relative abundance (No./Mb) | 75.28 | 64.63 | 65.18 | 60.86 | 52.64 |
| 密度 Relative density (bp/Mb) | 1 432.07 | 1 194.42 | 1 169.14 | 1 084.19 | 10 248.16 |
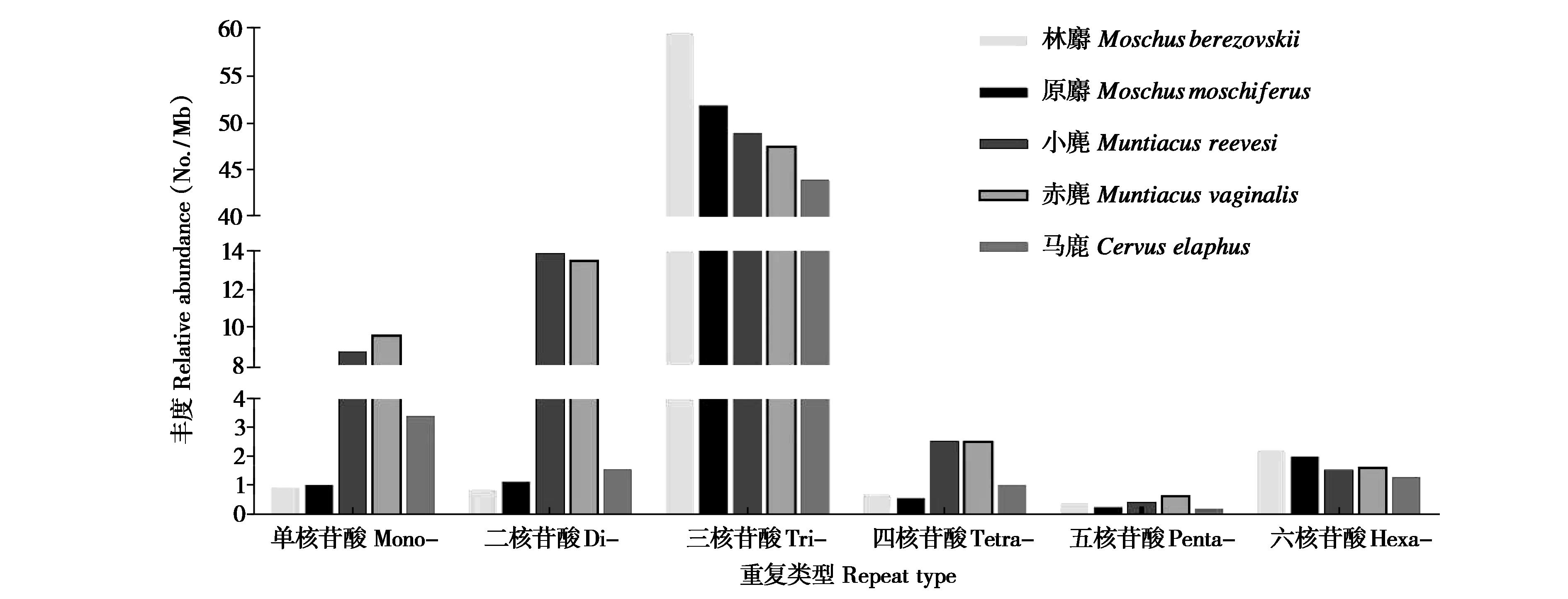
图1 林麝及其近缘物种蛋白质编码区不同重复拷贝类别SSRs丰度比较
Fig. 1 Comparison of SSR frequency of different repeat types in the CDS regions of the Moschus berezovskii and its closely related species
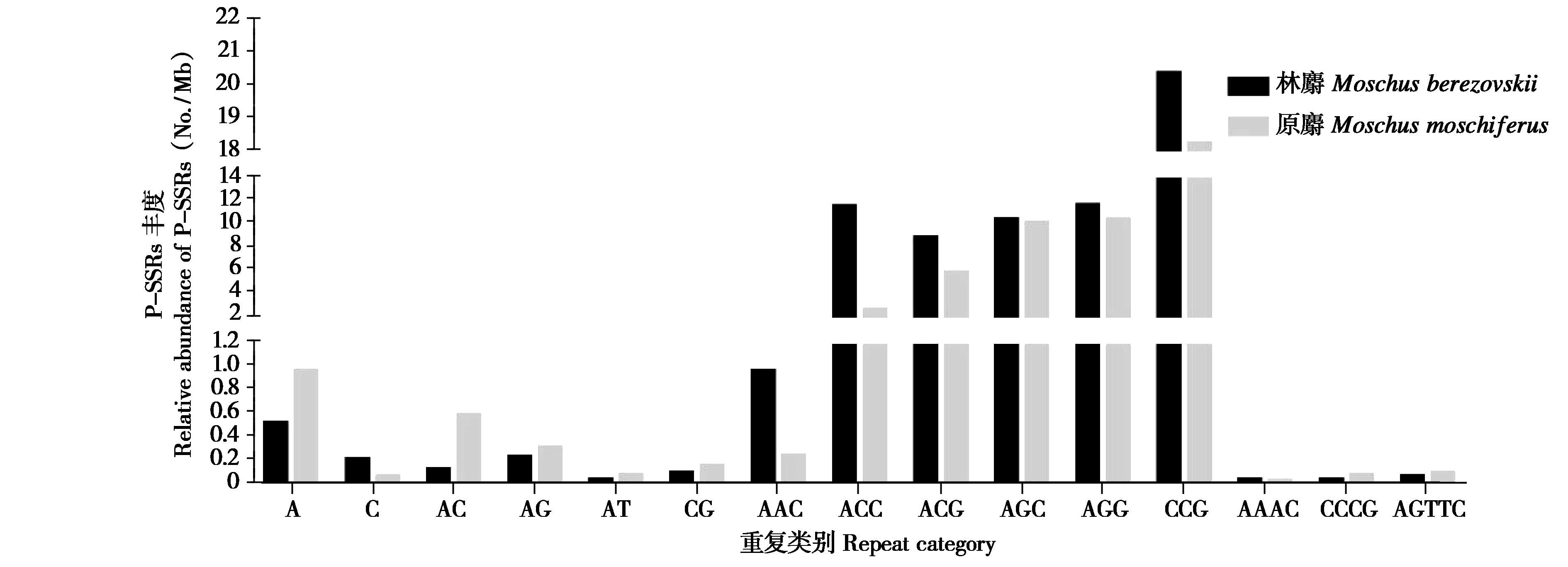
图2 林麝和原麝蛋白质编码区不同重复拷贝类别SSRs丰度比较
Fig. 2 Comparison of SSR frequency of different repeat category in the CDS regions of the Moschus berezovskii and Moschus moschiferus
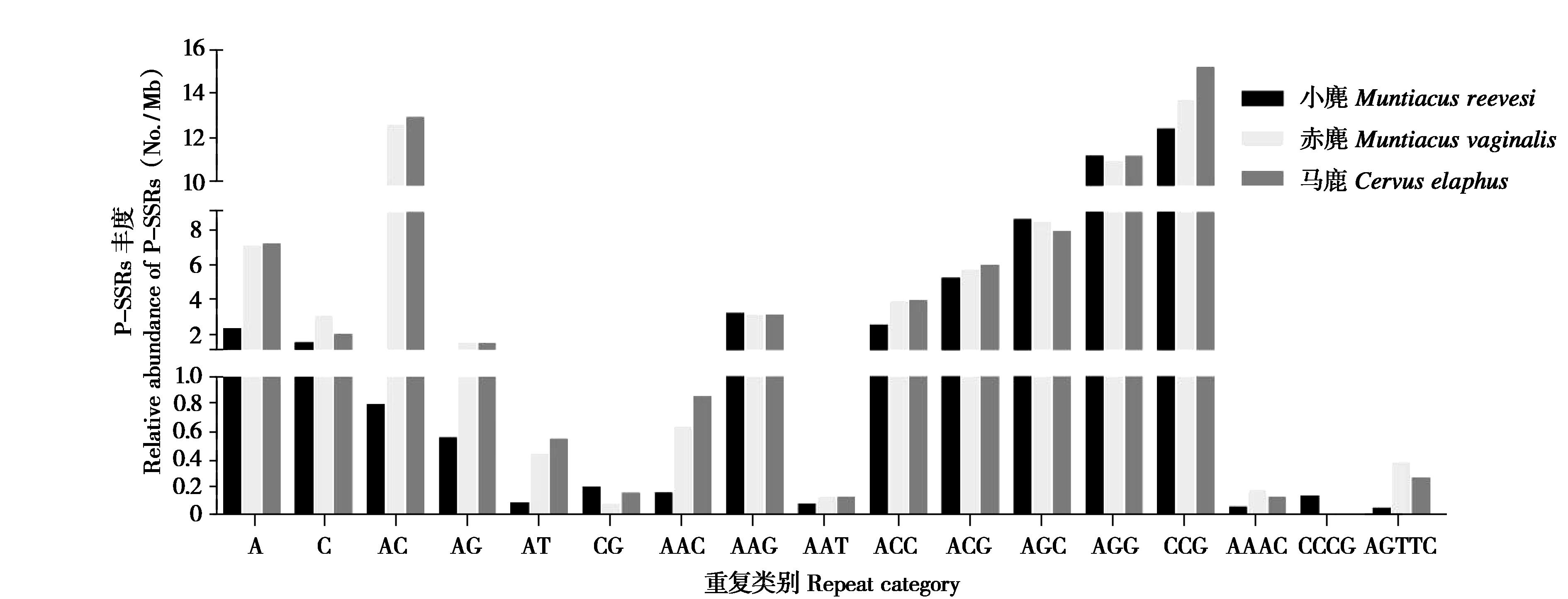
图3 马鹿、赤麂和小麂蛋白质编码区不同重复拷贝类别SSRs丰度比较
Fig. 3 Comparison of SSR frequency of different repeat category in the CDS regions of the Cervus elaphus, Muntiacus vaginalis and Muntiacus reeves
| GO term | 林麝 Moschus berezovskii | 原麝 Moschus moschiferus | 小麂 Muntiacus reeves | 赤麂 Muntiacus vaginalis | 马鹿 Cervus elaphus |
|---|---|---|---|---|---|
| Positive regulation of nitrogen compound metabolic process | + | + | + | + | + |
| Positive regulation of RNA biosynthetic process | + | + | + | + | + |
| Negative regulation of RNA biosynthetic process | + | + | + | + | - |
| Regulation of nucleobase-containing compound metabolic process | + | + | + | + | + |
| Regulation of nucleic acid-templated transcription | + | + | + | + | + |
| Transcription, DNA-templated | + | + | + | + | + |
| Regulation of biosynthetic process | + | + | + | + | + |
| Negative regulation of metabolic process | + | + | + | + | - |
| Positive regulation of metabolic process | + | + | + | + | + |
| Gene expression | + | + | + | + | + |
| Regulation of macromolecule biosynthetic process | + | + | + | + | + |
| Negative regulation of macromolecule biosynthetic process | + | + | + | + | - |
| Positive regulation of macromolecule metabolic process | + | + | + | + | + |
| Negative regulation of gene expression | + | + | - | + | - |
| Positive regulation of transcription, DNA-templated | + | + | + | + | + |
Negative regulation of nucleobase-containing compound metabolic process | + | + | - | + | - |
| Negative regulation of cellular metabolic process | + | + | - | + | - |
| Negative regulation of cellular biosynthetic process | + | + | + | + | - |
| Nucleic acid-templated transcription | + | + | - | + | + |
| Negative regulation of biological process | + | + | + | + | - |
| Stem cell differentiation | + | + | + | + | + |
| RNA biosynthetic process | + | + | - | + | + |
| Negative regulation of transcription by RNA polymerase Ⅱ | + | + | - | + | - |
| Regulation of nitrogen compound metabolic process | + | + | - | + | + |
| Regulation of RNA metabolic process | + | + | - | + | + |
| Central nervous system neuron differentiation | + | + | + | + | + |
| Negative regulation of RNA metabolic process | + | + | + | + | - |
| Regulation of RNA biosynthetic process | + | + | - | + | + |
| Negative regulation of transcription, DNA-templated | + | + | + | + | - |
| Positive regulation of cellular metabolic process | + | + | + | + | + |
表2 林麝及其近缘物种蛋白质编码区SSR序列的生物学过程的功能富集 (top 30)
Table 2 The most significantly enriched biological process GO terms of coding SSRs of Moschus berezovskii and its closely related species (top 30)
| GO term | 林麝 Moschus berezovskii | 原麝 Moschus moschiferus | 小麂 Muntiacus reeves | 赤麂 Muntiacus vaginalis | 马鹿 Cervus elaphus |
|---|---|---|---|---|---|
| Positive regulation of nitrogen compound metabolic process | + | + | + | + | + |
| Positive regulation of RNA biosynthetic process | + | + | + | + | + |
| Negative regulation of RNA biosynthetic process | + | + | + | + | - |
| Regulation of nucleobase-containing compound metabolic process | + | + | + | + | + |
| Regulation of nucleic acid-templated transcription | + | + | + | + | + |
| Transcription, DNA-templated | + | + | + | + | + |
| Regulation of biosynthetic process | + | + | + | + | + |
| Negative regulation of metabolic process | + | + | + | + | - |
| Positive regulation of metabolic process | + | + | + | + | + |
| Gene expression | + | + | + | + | + |
| Regulation of macromolecule biosynthetic process | + | + | + | + | + |
| Negative regulation of macromolecule biosynthetic process | + | + | + | + | - |
| Positive regulation of macromolecule metabolic process | + | + | + | + | + |
| Negative regulation of gene expression | + | + | - | + | - |
| Positive regulation of transcription, DNA-templated | + | + | + | + | + |
Negative regulation of nucleobase-containing compound metabolic process | + | + | - | + | - |
| Negative regulation of cellular metabolic process | + | + | - | + | - |
| Negative regulation of cellular biosynthetic process | + | + | + | + | - |
| Nucleic acid-templated transcription | + | + | - | + | + |
| Negative regulation of biological process | + | + | + | + | - |
| Stem cell differentiation | + | + | + | + | + |
| RNA biosynthetic process | + | + | - | + | + |
| Negative regulation of transcription by RNA polymerase Ⅱ | + | + | - | + | - |
| Regulation of nitrogen compound metabolic process | + | + | - | + | + |
| Regulation of RNA metabolic process | + | + | - | + | + |
| Central nervous system neuron differentiation | + | + | + | + | + |
| Negative regulation of RNA metabolic process | + | + | + | + | - |
| Regulation of RNA biosynthetic process | + | + | - | + | + |
| Negative regulation of transcription, DNA-templated | + | + | + | + | - |
| Positive regulation of cellular metabolic process | + | + | + | + | + |
Pathway Class | Pathway name | 林麝 Moschus berezovskii | 原麝 Moschus moschiferus | 小麂 Muntiacus reeves | 赤麂 Muntiacus muntjak | 马鹿 Cervus elaphus |
|---|---|---|---|---|---|---|
遗传信息调控 Genetic information processing | Protein families: genetic information processing | 1.83E-08 | 8.79E-14 | 1.55E-13 | 0 | 0 |
| Transcription factors | 9.35E-07 | 1.10E-13 | 1.33E-13 | 0 | 1.15E-11 | |
| Chromosome and associated proteins | 1.24E-05 | 1.76E-13 | 9.18E-04 | 6.37E-05 | 5.97E-09 | |
| Spliceosome | 4.03E-03 | 3.56E-06 | 1.15E-04 | 1.22E-05 | 1.26E-12 | |
| Transcription machinery | 2.36E-02 | 9.71E-04 | 1.32E-2 | 5.29E-3 | 1.64E-07 | |
| Ion channels | — | 5.50E-3 | 4.93E-3 | 5.93E-3 | 8.34E-07 | |
| Transcription | — | 1.82E-2 | 4.68E-2 | 4.46E-2 | 6.83E-3 | |
| Ribosome biogenesis | — | — | 4.10E-3 | 4.55E-3 | 3.73E-2 | |
新陈代谢 Metabolism | GnRH secretion | 3.78E-03 | 1.09E-05 | 1.80E-2 | — | 9.93E-3 |
| Lysine degradation | 8.37E-03 | 2.12E-07 | — | 4.53E-2 | 1.99E-3 | |
| Parathyroid hormone synthesis, secretion and action | — | 2.00E-2 | 3.08E-2 | — | — | |
| Renin secretion | — | 2.98E-2 | — | — | — | |
| Cortisol synthesis and secretion | — | — | 3.52E-2 | — | 3.59E-2 | |
| Insulin secretion | — | — | 3.83E-2 | — | 2.27E-2 | |
机体系统 Organismal systems | Dorso-ventral axis formation | — | 1.70E-03 | — | — | — |
| Endocrine system | — | 5.84E-3 | — | — | — | |
| Vascular smooth muscle contraction | — | 2.27E-2 | — | — | — | |
| Cell adherens junction | — | 2.19E-2 | — | — | 1.24E-2 | |
环境信息调控 Environmental information processing | Notch signaling pathway | 6.88E-03 | 9.53E-04 | 4.33E-3 | 6.37E-3 | 7.64E-05 |
| Signal transduction | 2.18E-02 | 1.25E-03 | — | — | 7.89E-3 | |
| cAMP signaling pathway | 4.33E-02 | — | — | — | — | |
| MAPK signaling pathway | — | 4.32E-03 | — | — | 2.08E-2 | |
| cGMP-PKG signaling pathway | — | 2.91E-2 | 4.01E-2 | — | 5.93E-3 | |
| Wnt signaling pathway | — | — | 2.85E-2 | — | — | |
疾病 Diseases | Maturity onset diabetes of the young | 1.63E-02 | 5.50E-03 | 3.63E-2 | 1.07E-2 | 2.21E-2 |
| Type Ⅱ diabetes mellitus | — | 1.78E-3 | — | 1.03E-2 | 1.02E-04 | |
| Bladder cancer | — | 2.04E-2 | — | — | 4.37E-2 | |
| Spinocerebellar ataxia | — | 2.05E-2 | — | — | — | |
| Breast cancer | — | 4.98E-2 | 2.28E-2 | — | — | |
| Cushing syndrome | — | — | 3.60E-3 | 5.97E-3 | 4.72E-2 |
表3 林麝及其近缘物种编码区SSR序列的KEGG通路富集 (top 30)
Table 3 The most significantly enriched KEGG pathway of coding SSRsof Moschus berezovskii and its closely related species (top 30)
Pathway Class | Pathway name | 林麝 Moschus berezovskii | 原麝 Moschus moschiferus | 小麂 Muntiacus reeves | 赤麂 Muntiacus muntjak | 马鹿 Cervus elaphus |
|---|---|---|---|---|---|---|
遗传信息调控 Genetic information processing | Protein families: genetic information processing | 1.83E-08 | 8.79E-14 | 1.55E-13 | 0 | 0 |
| Transcription factors | 9.35E-07 | 1.10E-13 | 1.33E-13 | 0 | 1.15E-11 | |
| Chromosome and associated proteins | 1.24E-05 | 1.76E-13 | 9.18E-04 | 6.37E-05 | 5.97E-09 | |
| Spliceosome | 4.03E-03 | 3.56E-06 | 1.15E-04 | 1.22E-05 | 1.26E-12 | |
| Transcription machinery | 2.36E-02 | 9.71E-04 | 1.32E-2 | 5.29E-3 | 1.64E-07 | |
| Ion channels | — | 5.50E-3 | 4.93E-3 | 5.93E-3 | 8.34E-07 | |
| Transcription | — | 1.82E-2 | 4.68E-2 | 4.46E-2 | 6.83E-3 | |
| Ribosome biogenesis | — | — | 4.10E-3 | 4.55E-3 | 3.73E-2 | |
新陈代谢 Metabolism | GnRH secretion | 3.78E-03 | 1.09E-05 | 1.80E-2 | — | 9.93E-3 |
| Lysine degradation | 8.37E-03 | 2.12E-07 | — | 4.53E-2 | 1.99E-3 | |
| Parathyroid hormone synthesis, secretion and action | — | 2.00E-2 | 3.08E-2 | — | — | |
| Renin secretion | — | 2.98E-2 | — | — | — | |
| Cortisol synthesis and secretion | — | — | 3.52E-2 | — | 3.59E-2 | |
| Insulin secretion | — | — | 3.83E-2 | — | 2.27E-2 | |
机体系统 Organismal systems | Dorso-ventral axis formation | — | 1.70E-03 | — | — | — |
| Endocrine system | — | 5.84E-3 | — | — | — | |
| Vascular smooth muscle contraction | — | 2.27E-2 | — | — | — | |
| Cell adherens junction | — | 2.19E-2 | — | — | 1.24E-2 | |
环境信息调控 Environmental information processing | Notch signaling pathway | 6.88E-03 | 9.53E-04 | 4.33E-3 | 6.37E-3 | 7.64E-05 |
| Signal transduction | 2.18E-02 | 1.25E-03 | — | — | 7.89E-3 | |
| cAMP signaling pathway | 4.33E-02 | — | — | — | — | |
| MAPK signaling pathway | — | 4.32E-03 | — | — | 2.08E-2 | |
| cGMP-PKG signaling pathway | — | 2.91E-2 | 4.01E-2 | — | 5.93E-3 | |
| Wnt signaling pathway | — | — | 2.85E-2 | — | — | |
疾病 Diseases | Maturity onset diabetes of the young | 1.63E-02 | 5.50E-03 | 3.63E-2 | 1.07E-2 | 2.21E-2 |
| Type Ⅱ diabetes mellitus | — | 1.78E-3 | — | 1.03E-2 | 1.02E-04 | |
| Bladder cancer | — | 2.04E-2 | — | — | 4.37E-2 | |
| Spinocerebellar ataxia | — | 2.05E-2 | — | — | — | |
| Breast cancer | — | 4.98E-2 | 2.28E-2 | — | — | |
| Cushing syndrome | — | — | 3.60E-3 | 5.97E-3 | 4.72E-2 |
| 基因名称 | 微卫星类型 | Map ID | KEGG pathway |
|---|---|---|---|
| CEBPB | (GCC)5 (GCC)8 | Map04657 Map05152 Map05202 | IL-17 signaling pathway Tuberculosis Transcriptional misregulation in cancer |
| ZEB1 | (GAG)5 | Map05206 Map05202 Map05215 | MicroRNAs in cancer Transcriptional misregulation in cancer Prostate cancer |
| THY1 | (GCT)6 | Map04670 | Leukocyte transendothelial migration |
| Bcl2 | (CCG)6 | Map05200 Map05206 Map05022 Map05202 Map04115 Map05210 Map05212 Map05220 Map05222 Map05215 Map05226 Map04932 Map05012 Map05221 Map05225 | Pathways in cancer MicroRNAs in cancer Pathways of neurodegeneration-multiple diseases Transcriptional misregulation in cancer p53 signaling pathway Colorectal cancer Pancreatic cancer Chronic myeloid leukemia Small cell lung cancer Prostate cancer Gastric cancer Non-alcoholic fatty liver disease Parkinson disease Acute myeloid leukemia Hepatocellular carcinoma |
| PDCD1 | (CTG)6 | Map04660 Map05235 | T cell receptor signaling pathway PD-L1 expression and PD-1 checkpoint pathway in cancer |
| WAS | (ATG)6 | Map04933 Map04936 Map05231 | AGE-RAGE signaling pathway in diabetic complications Alcoholic liver disease Choline metabolism in cancer |
| HDAC5 | (AGC)5 (GAC)5 (GGA)5 | Map05203 Map05206 | Viral carcinogenesis MicroRNAs in cancer |
| MYH9 | (GGA)5 | Map05130 | Pathogenic Escherichia coli infection |
| JAG2 | (GCT)5 | Map05200 Map05224 | Pathways in cancer Breast cancer |
| NOTCH4 | (GCT)5 | Map05206 Map05224 Map05200 | MicroRNAs in cancer Breast cancer Pathways in cancer |
表4 林麝关键免疫基因所含SSR类型及其KEGG 通路
Table 4 SSR types of key immune genes in Moschus berezovskii and their KEGG pathways
| 基因名称 | 微卫星类型 | Map ID | KEGG pathway |
|---|---|---|---|
| CEBPB | (GCC)5 (GCC)8 | Map04657 Map05152 Map05202 | IL-17 signaling pathway Tuberculosis Transcriptional misregulation in cancer |
| ZEB1 | (GAG)5 | Map05206 Map05202 Map05215 | MicroRNAs in cancer Transcriptional misregulation in cancer Prostate cancer |
| THY1 | (GCT)6 | Map04670 | Leukocyte transendothelial migration |
| Bcl2 | (CCG)6 | Map05200 Map05206 Map05022 Map05202 Map04115 Map05210 Map05212 Map05220 Map05222 Map05215 Map05226 Map04932 Map05012 Map05221 Map05225 | Pathways in cancer MicroRNAs in cancer Pathways of neurodegeneration-multiple diseases Transcriptional misregulation in cancer p53 signaling pathway Colorectal cancer Pancreatic cancer Chronic myeloid leukemia Small cell lung cancer Prostate cancer Gastric cancer Non-alcoholic fatty liver disease Parkinson disease Acute myeloid leukemia Hepatocellular carcinoma |
| PDCD1 | (CTG)6 | Map04660 Map05235 | T cell receptor signaling pathway PD-L1 expression and PD-1 checkpoint pathway in cancer |
| WAS | (ATG)6 | Map04933 Map04936 Map05231 | AGE-RAGE signaling pathway in diabetic complications Alcoholic liver disease Choline metabolism in cancer |
| HDAC5 | (AGC)5 (GAC)5 (GGA)5 | Map05203 Map05206 | Viral carcinogenesis MicroRNAs in cancer |
| MYH9 | (GGA)5 | Map05130 | Pathogenic Escherichia coli infection |
| JAG2 | (GCT)5 | Map05200 Map05224 | Pathways in cancer Breast cancer |
| NOTCH4 | (GCT)5 | Map05206 Map05224 Map05200 | MicroRNAs in cancer Breast cancer Pathways in cancer |
| Bana N Á, Nyiri A, Nagy J, Frank K, Nagy T, Stéger V, Schiller M, Lakatos P, Sugár L, Horn P, Barta E, Orosz L. 2018. The red deer Cervus elaphus genome CerEla1.0: sequencing, annotating, genes, and chromosomes. Molecular Genetics and Genomics, 293 (3): 665-684. | |
| Chen C J, Chen H, Zhang Y, Thomas H R, Margaret H F, He Y H, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Molecular Plant, 13 (8): 1194-1202. | |
| Du L, Li Y, Zhang X, Yue B. 2013. A user‑friendly program for reporting distribution and building databases of microsatellites from genome sequences. The Journal of Heredity, 104: 154-157. | |
| Du L M, Zhang C, Liu Q, Zhang X Y, Yue B S. 2018. Krait: an ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics, 34 (4): 681-683. | |
| Duval A, Hamelin R. 2002. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Research, 62 (9): 2447-2454. | |
| Duval A, Gayet J, Zhou X P, Iacopetta B, Thomas G, Hamelin R. 1999. Frequent frameshift mutations of the TCF-4 gene in colorectal cancers with microsatellite instability. Cancer Research, 59 (17): 4213-4215. | |
| Fan Z X, Li W J, Jin J, Cui K, Yan C C, Peng C J, Jian Z Y, Bu P, Price M, Zhang X Y, Shen Y M, Li J, Qi W H, Yue B S. 2018. The draft genome sequence of forest musk deer (Moschus berezovskii). Giga Science, 7 (4): giy038. | |
| Jiang X M, Hu T Z, Xiang X S, Qi W H. 2015. Statistics and bioinformatics analysis of microsatellite sequence in poplar whole genomes. Southwest China Journal of Agricultural Sciences, 28 (2): 527-533. (in Chinese) | |
| Jin P, Warren S T. 2000. Understanding the molecular basis of fragile X syndrome. Human Molecular Ggenetics, 9 (6): 901-908. | |
| Kashi Y, King D, Soller M. 1997. Simple sequence repeats as a source of quantitative genetic variation. Trends in Genetics, 13 (2): 74-78. | |
| Li W J, Li Y Z, Du L M, Huang J, Shen Y M, Zhang X Y, Yue B S. 2014. Comparative analysis of microsatellite sequences distribution in the genome of giant panda and polar bear. Sichuan Journal of Zoology, 33 (6): 874-878 (in Chinese) | |
| Li Y C. 2004. Microsatellites within genes: structure, function, and evolution. Molecular Biology and Evolution, 21 (6): 991-1007. | |
| Lu T, Wang C, Du C, Liu S, Shen Y M, Zhang X Y, Yue B S. 2017. Distribution regularity of microsatellites in Moschus berezovskii genome. Sichuan Journal of Zoology, 36 (4): 420-424. (in Chinese) | |
| Metzgar D, Wills C. 2000. Evidence for the adaptive evolution of mutation rates. Cell, 101 (6): 581-584. | |
| Montgelard C, Catzeflis F, Douzery E. 1997. Phylogenetic relationships among cetartiodactyls andcetaceans as deduced from the comparison of cytochrome b and 12S RNA mitochondrial sequences. Molecular Biology Evolution, 14: 550-559. | |
| O’Dushlaine C T, Edwards R J, Park S D, et al. 2005. Tandem repeat copy-number variation in protein-coding regions of human genes. Genome Biology, 6 (8): R69. | |
| Ohtaishi N, Gao Y. 1990. A review of the distribution of all species of deer (Tragulidae, Moschidae and Cervidae) in China. Mammal Review, 20 (2-3): 125-144. | |
| Qi W H, Jiang X M, Xiao G S, Huang X Y, Du L M. 2013. Seeking and bioinformatics analysis of microsatellite sequence in the genomes of cow and sheep. Acta Veterinaria et Zootechnica Sinica, 44 (11): 1724-1733. (in Chinese) | |
| Qi W H, Lu T, Zheng C L, Jiang X M, Jie H, Zhang X Y, Yue B S, Zhao G J. 2020.Distribution patterns of microsatellites and development of its marker in different genomic regions of forest musk deer genome based on high throughput sequencing. Aging, 12 (5): 4445-4462. | |
| Sermon K, Seneca S, Rycke M D, Goossens V, Van de Velde H, De Vos A, Platteau P, Lissens W, Van Steirteghem A, Liebaers I. 2001. PGD in the lab for triplet repeat diseases‒myotonic dystrophy, Huntington’s disease and Fragile‒X syndrome. Molecular and Cellular Endocrinology, 183: S77-S85. | |
| Schwartz S, Yamamoto H, Navarro M, Maestro M, Reventós J, Perucho M. 1999. Frameshift mutations at mononucleotide repeats in caspase - 5 and other target genes in endometrial and gastrointestinal cancer of the microsatellite mutator phenotype. Cancer Research, 59 (12): 2995-3002. | |
| Subramanian S, Mishra RK, Singh L. 2003. Genome‑wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biology, 4 (2): 1-10. | |
| Timchenko N A, Iakova P, Cai Z J, Smith J R, Timchenko L T. 2001. Molecular basis for impaired muscle differentiation in myotonic dystrophy. Molecular and Cellular Biology, 21 (20): 6927-6938. | |
| Vassileva V, Millar A, Briollais L, Chapman W, Bapat B. 2002. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Research, 62 (14): 4095-4099. | |
| Wang X H, Pan X F, Li H Q, Duan F. 2016. Advances in the studies of the expansion of (CAG)n·(CTG)n trinucleotide repeats and mecha-nisms underlying its related diseases. International Journal of Genetics, 39 (5): 274-281. (in Chinese) | |
| Wang Z L, Huang J, Du L M, Li W J, Yue B S, Zhang X Y. 2013. Comparison of microsatellites between the genomes of Tetranychus urticae and Ixodes scapularis . Sichuan Journal of Zoology, 32 (4): 481-486. (in Chinese) | |
| Yi L, Dalai M G, Su R N, Lin W L, Erdenedalai M, Luvsantseren B, Chimedtseren C, Wang Z, Hasi S R. 2020. Whole‑genome sequencing of wild Siberian musk deer (Moschus moschiferus) provides insights into its genetic features. BMC Genomics, 21(1): 108. | |
| Yin Y, Fan H Z, Zhou B T, Hu Y B, Fan G Y, Wang J H, Zhou F, Nie W H, Zhang C Z, Liu L, Zhong Z Y, Zhu W B, Liu G C, Lin Z S, Liu C, Zhou J, Huang G P, Li Z H, Yu J P, Zhang Y L, Yang Y, Zhuo B Z, Zhang B W, Chang J, Qian H Y, Peng Y M, Chen X Q, Chen L, Li Z P, Zhou Q, Wang W, Wei F W. 2021. Molecular mechanisms and topological consequences of drastic chromosomal rearrangements of muntjac deer. Nature Communication, 12 (1): 6858. | |
| Yuki M, Masumi I, Shujiro O, Yoshizawa A C, Minoru K. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Research, 35 (Web Server issue): W182-W185. | |
| 丁戈, 姚南, 吴琼, 刘恒, 郑国锠. 2008. 着丝粒结构与功能研究的新进展. 植物学通报, 25 (2): 149-160. | |
| 王希恒, 潘学峰, 李红权, 段斐. 2016. (CAG)n·(CTG)n三核苷酸重复序列扩增及相关疾病机制研究进展. 国际遗传学杂志, 39 (5): 274-281. | |
| 卢婷, 王晨, 杜超, 刘姝, 沈咏梅, 张修月, 岳碧松. 2017. 林麝全基因组微卫星分布规律研究. 四川动物, 36 (4): 420-424. | |
| 刘志霄, 盛和林. 2000. 我国麝的生态研究与保护问题概述. 动物学杂志, 35 (3): 54-57. | |
| 李午佼, 李玉芝, 杜联明, 黄杰, 沈咏梅, 张修月, 岳碧松. 2014. 大熊猫和北极熊基因组微卫星分布特征比较分析. 四川动物, 33 (6): 874-878. | |
| 杨述林, 王志刚, 樊斌, 刘榜, 李奎. 2003. 微卫星及其功能研究. 湖北农业科学, 2003 (6): 91-93. | |
| 汪自立, 黄杰, 杜联明, 李午佼, 岳碧松, 张修月. 2013. 二斑叶螨和肩突硬蜱基因组微卫星分布规律研究. 四川动物, 32 (4): 481-486. | |
| 戚文华, 蒋雪梅,肖国生, 黄小云, 杜联明. 2013. 牛和绵羊全基因组微卫星序列的搜索及其生物信息学分析. 畜牧兽医学报, 44 (11): 1724-1733. | |
| 蒋雪梅, 胡廷章, 向兴胜, 戚文华. 2015. 杨树全基因组微卫星序列的统计及其生物信息学分析. 西南农业学报, 28 (2): 527-533. |
| [1] | 卢文代, 陆娇, 雷苑. 云南糯扎渡省级自然保护区发现黑化赤麂[J]. 兽类学报, 2024, 44(3): 394-394. |
| [2] | 余苗杰, 徐忠鲜, 蒋雪梅, 王春花, 赵婵娟, 戚文华, 竭航. 林麝不同组织miRNA表达谱[J]. 兽类学报, 2023, 43(6): 723-733. |
| [3] | 邵雪峰, 平晓莹, 李月圆, 陈龙, 聂志文, 胡远满, 李月辉. 小兴安岭马鹿粪便的分解速率[J]. 兽类学报, 2023, 43(2): 157-163. |
| [4] | 江峰, 宋鹏飞, 张婧捷, 高红梅, 汪海静, 蔡振媛, 刘道鑫, 张同作. 不同养殖场林麝肠道微生物组成和功能的差异[J]. 兽类学报, 2023, 43(2): 129-140. |
| [5] | 田新民, 张明海. 基于粪便DNA的东北马鹿遗传多样性[J]. 兽类学报, 2023, 43(1): 41-49. |
| [6] | 胡娟, 谢培根, 李婷婷, 郭瑞, 许丽娟, 宋虓, 徐爱春. 同域分布的黑麂和小麂的时空生态位分化[J]. 兽类学报, 2022, 42(6): 641-651. |
| [7] | 周良俊, 王琳, 魏楷丽, 张明海, 张玮琪. 内蒙古森林—草原交错带中野生东北马鹿冬季食物组成模式[J]. 兽类学报, 2022, 42(3): 240-249. |
| [8] | 张沼, 张蕊, 李晓宇, 赛罕, 杨振东, 韩志庆, 鲍伟东. 内蒙古赛罕乌拉自然保护区东北马鹿种群遗传多样性与性别结构分析[J]. 兽类学报, 2021, 41(1): 42-50. |
| [9] | 周虎, 刘周, 庞春梅, 陈康民, 章书声, 杨淑贞, 徐爱春. 浙江天目山国家级自然保护区三种鹿科动物舔盐行为比较[J]. 兽类学报, 2021, 41(1): 99-107. |
| [10] | 高惠 乔付杰 滕丽微 李俊乐 余梦琦 刘振生. 马鹿阿拉善亚种遗传多样性和群体结构[J]. 兽类学报, 2020, 40(5): 458-466. |
| [11] | 王静 白瑞丹 蔡永华 黎勇 程建国 付文龙 周密 盛岩 孟秀祥. 圈养林麝麝香分泌时间节律及影响因素[J]. 兽类学报, 2020, 40(5): 485-492. |
| [12] | 刘嘉辉 王艳 边坤 唐婕 王伟峰 郭林文 王波 方谷 赵兰 齐晓光. 重引入林麝的家域利用与个体迁移[J]. 兽类学报, 2020, 40(2): 109-119. |
| [13] | 王豆 许冠 王洪永 何森 卜书海 郑雪莉. 中国圈养林麝微卫星DNA多样性研究[J]. 兽类学报, 2019, 39(6): 599-607. |
| [14] | 罗娟娟 秦家慧 李佳琦 兰广成 郭志宏 徐永恒 宋森. 宁夏兽类新纪录——小麂 (Muntiacus reevesi Ogilby, 1839)[J]. 兽类学报, 2019, 39(6): 688-693. |
| [15] | 房璇 孙太福 蔡永华 董霞 黎勇 周密 孟秀祥. 圈养林麝的活跃性及与社会亲和度的关系[J]. 兽类学报, 2019, 39(5): 531-536. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2