

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2023, Vol. 43 ›› Issue (2): 129-140.DOI: 10.16829/j.slxb.150701
• 研究论文 • 下一篇
江峰1,3( ), 宋鹏飞1,2,3, 张婧捷1,2,3, 高红梅1,3, 汪海静1,2,3, 蔡振媛1,3, 刘道鑫1,2,3, 张同作1,3(
), 宋鹏飞1,2,3, 张婧捷1,2,3, 高红梅1,3, 汪海静1,2,3, 蔡振媛1,3, 刘道鑫1,2,3, 张同作1,3( )
)
收稿日期:2022-06-06
接受日期:2022-10-20
出版日期:2023-03-30
发布日期:2023-03-23
通讯作者:
张同作
作者简介:江峰 (1992- ),男,博士,主要从事动物生态学、保护生物学研究. E-mail: jiangfeng@nwipb.cas.cn
基金资助:
Feng JIANG1,3( ), Pengfei SONG1,2,3, Jingjie ZHANG1,2,3, Hongmei GAO1,3, Haijing WANG1,2,3, Zhenyuan CAI1,3, Daoxin LIU1,2,3, Tongzuo ZHANG1,3(
), Pengfei SONG1,2,3, Jingjie ZHANG1,2,3, Hongmei GAO1,3, Haijing WANG1,2,3, Zhenyuan CAI1,3, Daoxin LIU1,2,3, Tongzuo ZHANG1,3( )
)
Received:2022-06-06
Accepted:2022-10-20
Online:2023-03-30
Published:2023-03-23
Contact:
Tongzuo ZHANG
摘要:
肠道疾病是养殖林麝 (Moschus berezovskii) 常见疾病。动物肠道微生物伴随宿主进化并与胃肠道构成了复杂的微生态系统。为探究不同饲养环境对圈养林麝肠道微生物组成和功能的影响,本研究对采自国内5个不同养殖场的215份粪便样品进行了16S rRNA基因高通量测序,对比分析不同养殖场林麝肠道微生物组成、多样性和功能的差异。结果显示,厚壁菌门和拟杆菌门是未喂食复合益生菌的祁连县养殖场林麝肠道菌群的绝对优势菌门,而喂食复合益生菌的甘肃两当县和陕西凤县的4家养殖场林麝肠道菌群的绝对优势菌门为厚壁菌门和变形菌门。不同养殖场林麝肠道菌群组成、优势菌门、优势菌属、潜在致病菌、代谢及疾病相关功能均有显著差异。祁连县养殖场林麝肠道微生物的α多样性和疾病相关功能表达量显著低于其他养殖场,并以肠型2为主,其主导菌为厚壁菌门、UCG-005和拟杆菌属;两当县和凤县的4家养殖场林麝肠道菌群潜在致病菌相对丰度较低。本研究推测食物组成差异可能是导致不同养殖场林麝肠道微生物差异的主要因素,复合益生菌的使用可能是导致α多样性和潜在致病菌下降的重要因素。该结果可为林麝的人工养殖和有效管理提供科学依据,也对人工饲养环境评估和未来的再引入计划具有一定指导意义。
中图分类号:
江峰, 宋鹏飞, 张婧捷, 高红梅, 汪海静, 蔡振媛, 刘道鑫, 张同作. 不同养殖场林麝肠道微生物组成和功能的差异[J]. 兽类学报, 2023, 43(2): 129-140.
Feng JIANG, Pengfei SONG, Jingjie ZHANG, Hongmei GAO, Haijing WANG, Zhenyuan CAI, Daoxin LIU, Tongzuo ZHANG. Comparative analysis of gut microbial composition and functions of forest musk deer in different breeding centres[J]. ACTA THERIOLOGICA SINICA, 2023, 43(2): 129-140.
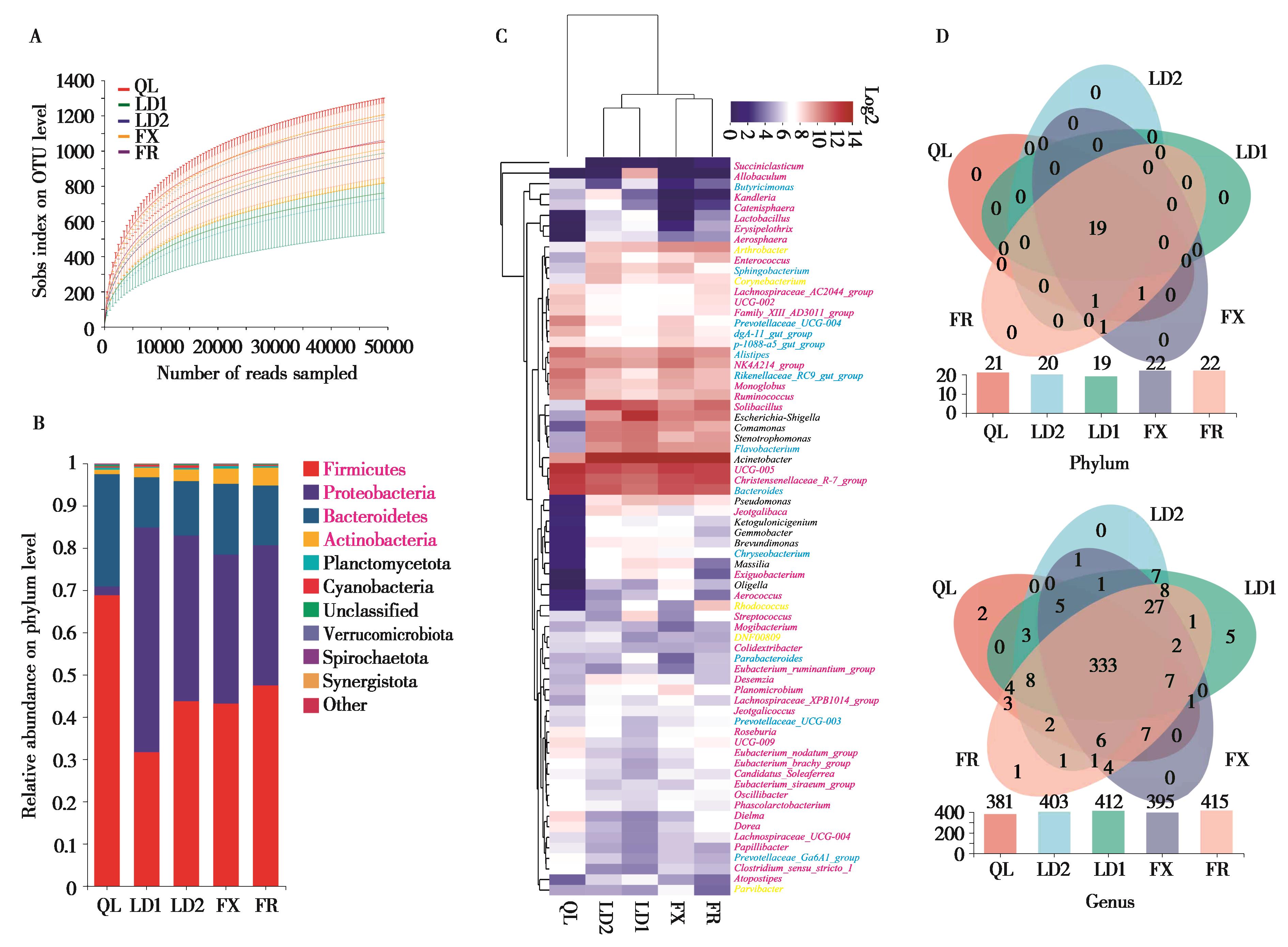
图1 不同养殖场林麝肠道微生物组成分析. A:基于Sobs指数的测序样本稀释曲线;B:门相对丰度柱状图,红色字体表示优势菌门;C:可鉴别细菌属 (Top 70) 聚类热图分析,红色、蓝色、黑色、橙色和绿色文字分别表示厚壁菌门、拟杆菌门、变形菌门、放线菌门和浮霉菌门的细菌属;D:门和属水平Venn图分析. QL:祁连县养殖场;LD:两当县养殖场;FX:凤县养殖场;FR:散养场. 以下图中注释相同
Fig. 1 Analyses of gut microbial composition of forest musk deers (FMD) in different breeding centres. A: Rarefaction curves based on Sobs; B: Relative abundance of bacterial phyla. The dominant phyla were shown in red; C: Cluster heat map analysis of identifiable bacterial genera (Top 70), the red, blue, black, orange and green text indicated Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and Planctomycetes, respectively. D: Venn diagrams at phylum and genus level. QL: Farm in Qilian County; LD: Farm in Liangdang County; FX: Farm in Feng County; FR:Free-range farm. Same notes in the following figure
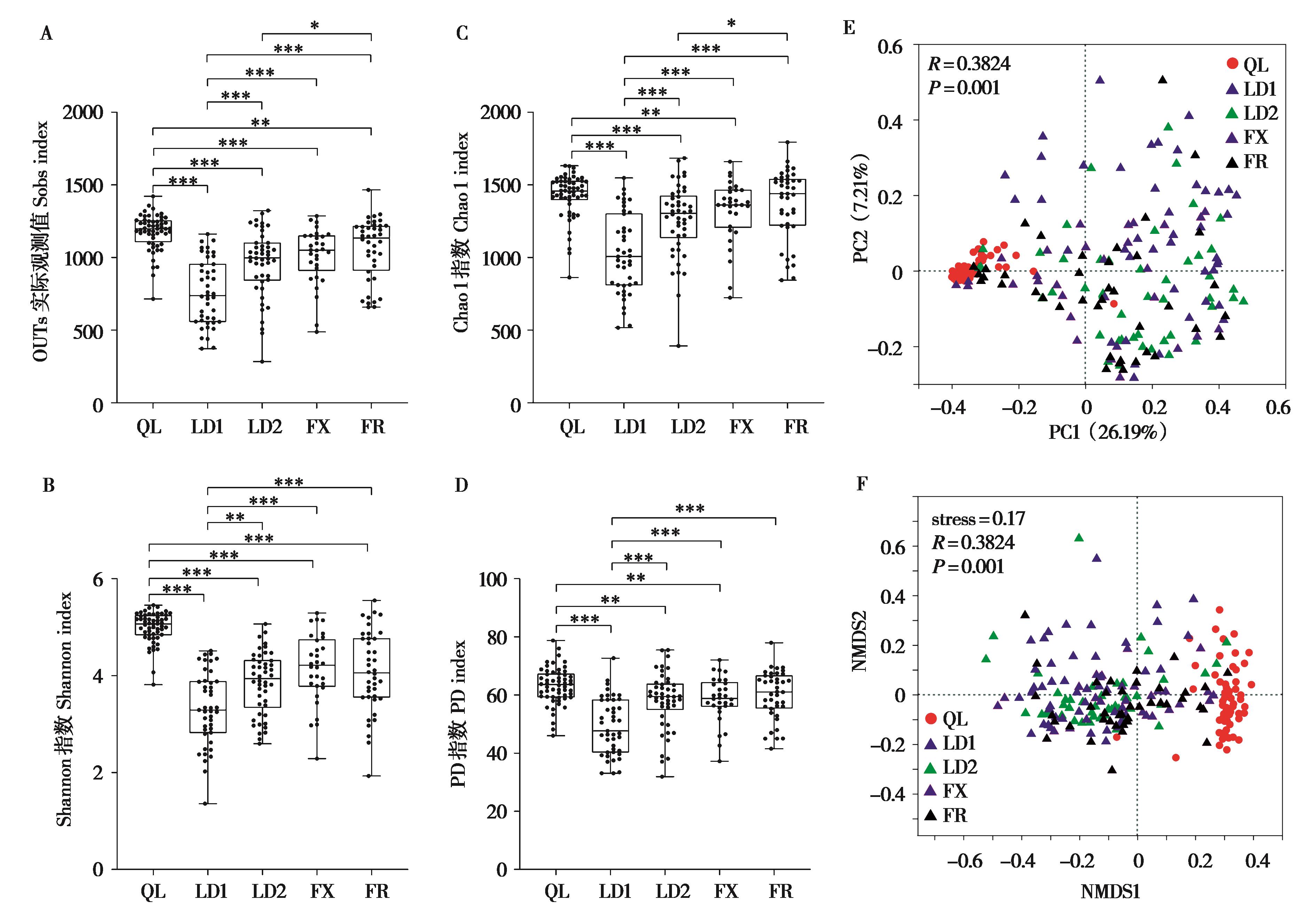
图2 不同养殖场林麝肠道微生物多样性分析. Sobs (A)、Shannon (B)、Chao1 (C) 和 PD (D) 指数的α多样性组间差异分析;不同养殖场林麝肠道菌群组间PCoA聚类分析 (E) 和NMDS聚类分析 (F). *P < 0.05 (Wilcoxon rank-sum test),**P < 0.01,***P < 0.001
Fig. 2 Analyses of gut microbial diversity of forest musk deers in different breeding centres. Analysis of differences in α diversity between groups based on Sobs (A), Shannon (B), Chao1 (C) and PD (D) indices. PCoA analysis (E) and NMDS analysis (F) among different farms. *P < 0.05 (Wilcoxon rank-sum test), **P < 0.01, ***P < 0.001
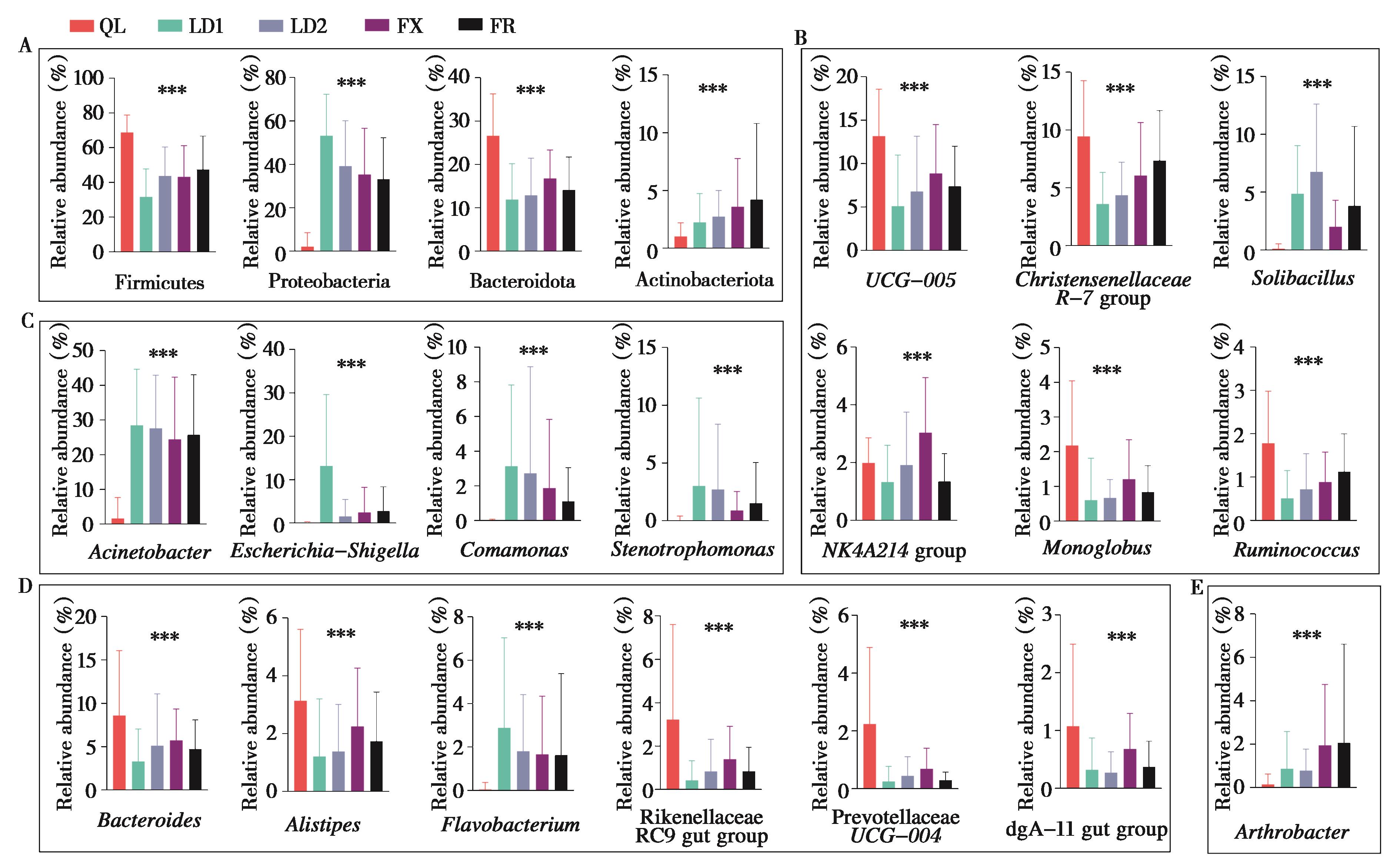
图3 不同养殖场林麝肠道菌群优势菌组间差异分析. 优势菌门组间差异分析 (A);厚壁菌门 (B)、变形菌门 (C)、拟杆菌门 (D) 和放线菌门 (E) 中优势菌属组间差异分析. ***P < 0.001 (Kruskal-Wallis test)
Fig. 3 Differences of dominant bacteria in gut microbiota of forest musk deers in different breeding centres. Analysis of the differences between the dominant phyla (A); Analysis of the differences between the dominant genera belonging to Firmicutes (B), Proteobacteria (C), Bacteroidetes (D), Actinobacteria (E). ***P < 0.001 (Kruskal-Wallis test)
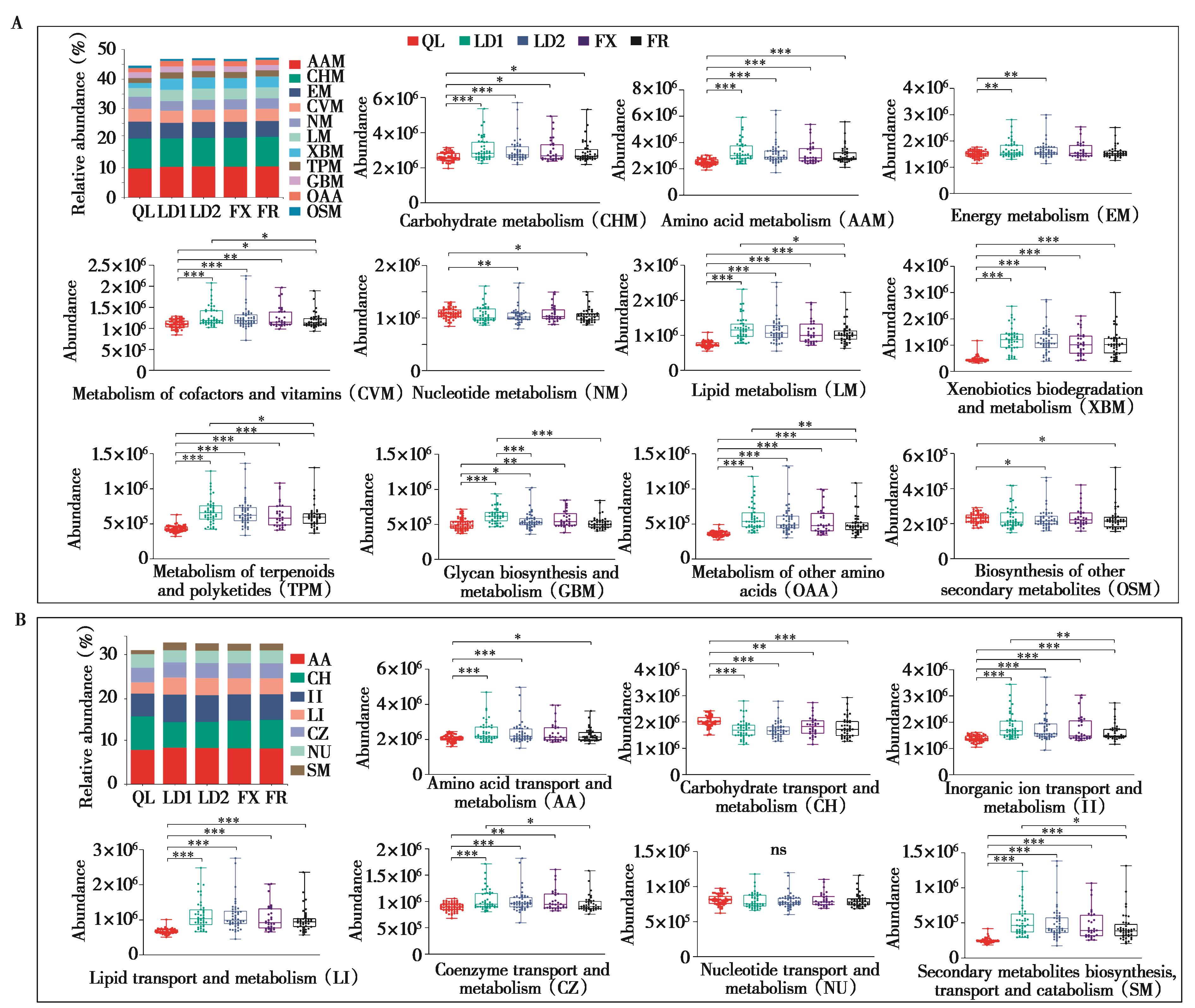
图4 基于KEGG数据库 (A) 和eggNOG数据库 (B) 的功能注释及level 2水平上代谢功能的组间差异分析. *P < 0.05 (Wilcoxon rank-sum test),**P < 0.01,***P < 0.001. ns,不显著
Fig. 4 Functional annotation based on KEGG database (A) and eggNOG database (B), and analysis of the differences of metabolic functions between groups at the level-2 level. **P < 0.01, ***P < 0.001. ns, not significant

图5 不同养殖场林麝肠道菌群中潜在致病菌 (A) 与疾病相关功能富集 (B) 组间差异分析. *P < 0.05,**P < 0.01,***P < 0.001,ns,无显著差异
Fig. 5 Analysis of the differences of potential pathogenic bacteria (A) and disease related functions (B) of forest musk deers in different breeding centres. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant
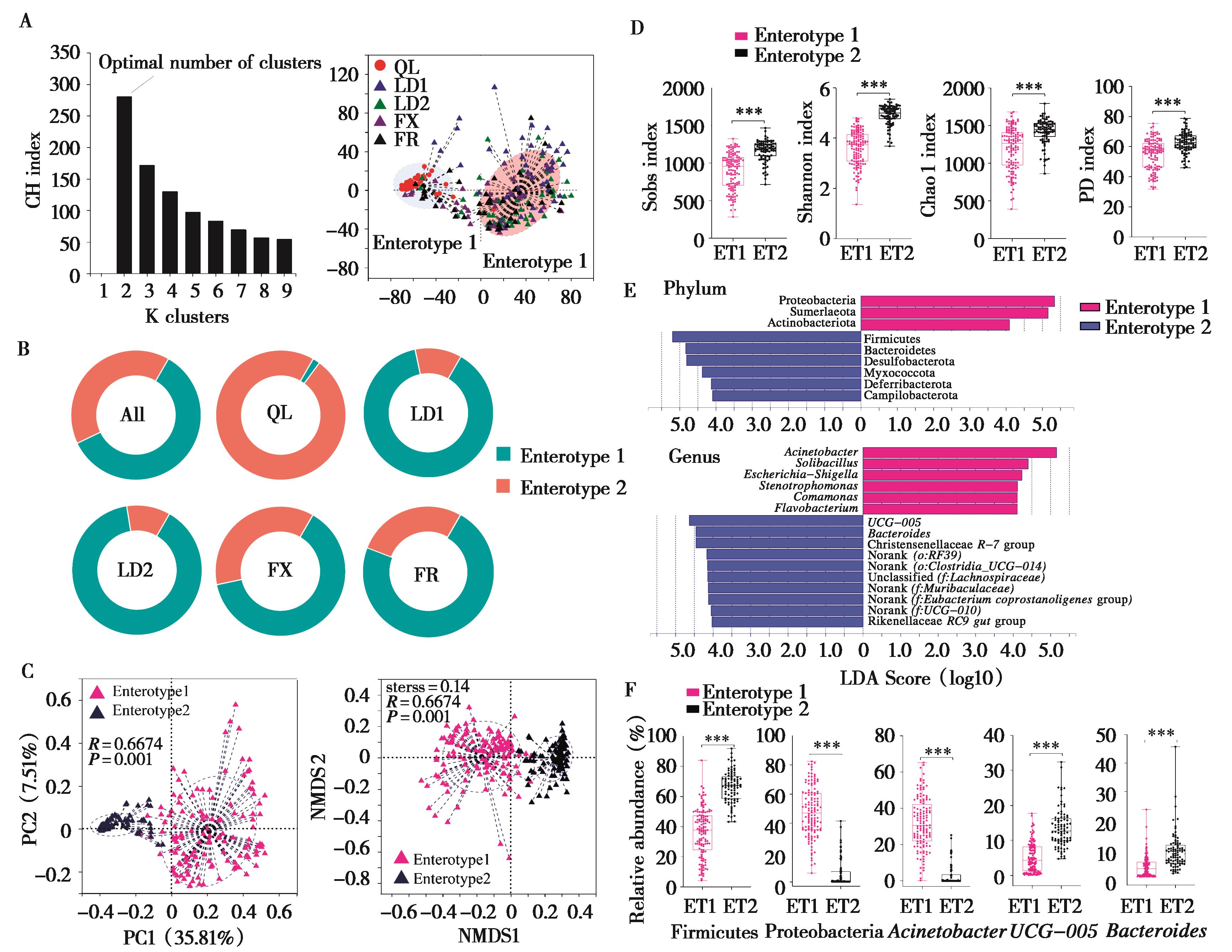
图6 不同养殖场林麝肠型差异分析. A:最优集群数分析和肠型分型图分析;B:不同养殖场林麝肠道菌群肠型分布分析;C:不同肠型α多样性差异性分析;D:门水平和属水平上不同肠型LEfSe分析;E:不同肠型生物标记物 (biomarker) 组间差异分析
Fig. 6 Analysis of different enterotype in forest musk deers from different breeding centres. A: Analysis of optimal cluster number and enterotype map; B: Distribution of enterotype in different farms; C: Analysis of α diversity in different enterotypes; D: LEfSe analysis of different enterotypes at phylum and genus levels; E: Difference analysis of biomarkers of different enterotypes
| Bäckhed F, Ley R E, Sonnenburg J L, Peterson D A, Gordon J I. 2005. Host-bacterial mutualism in the human intestine. Science, 307 (5717): 1915-1920. | |
| Bolger A M, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics, 30: 2114-2120. | |
| Chen L, Liu M, Zhu J, Gao Y, Sha W L, Ding H X, Jiang W J, Wu S P. 2020. Age, gender, and feeding environment influence fecal microbial diversity in Spotted Hyenas (Crocuta crocuta). Current Microbiology, 77 (7): 1139-1149. | |
| Costello E K, Lauber C L, Hamady M, Fierer N, Gordon J I, Knight R. 2009. Bacterial community variation in human body habitats across space and time. Science, 326 (5960): 1694-1697. | |
| Cui X X, Wang Z F, Yan T H, Chang S H, Wang H, Hou F J. 2019. Rumen bacterial diversity of Tibetan sheep (Ovis aries) associated with different forage types on the Qinghai-Tibetan Plateau. Canadian Journal of Microbiology, 65 (12): 859-869. | |
| David L A, Maurice C F, Carmody R N, Gootenberg D B, Button J E, Wolfe B E, Ling A V, Devlin A S, Varma Y, Fischbach M A, Biddinger S B, Dutton R J, Turnbaugh P J. 2014. Diet rapidly and reproducibly alters the human gut microbiome. Nature, 505 (7484): 559-563. | |
| De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet J B, Massart S, Collini S, Pieraccini G, Lionetti P. 2010. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of The National Academy of Sciences of The United States of America, 107 (33): 14691-14696. | |
| Edgar R C. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10 (10): 996-998. | |
| Ezenwa V O, Gerardo N M, Inouye D W, Medina M, Xavier J B. 2012. Microbiology. Animal behavior and the microbiome. Science, 338 (6104): 198-199. | |
| Fan J M, Zheng X L, Wang H Y, Qi H, Jiang B M, Qiao M P, Zhou J W, Bu S H. 2019. Analysis of genetic diversity and population structure in three forest musk deer captive populations with different origins. G3-Genes Genomes Genetics, 9 (4): 1037-1044. | |
| Fan Z X, Li W J, Jin J Z, Cui K, Yan C C, Peng C J, Jian Z Y, Bu P, Price M, Zhang X Y, Shen Y M, Li J, Qi W H, Yue B S. 2018. The draft genome sequence of forest musk deer (Moschus berezovskii). Gigascience, 7 (4): 1-6. | |
| Fountain‑Jones N M, Clark N J, Kinsley A C, Carstensen M, Forester J, Johnson T J, Miller E A, Moore S, Wolf T M, Craft M E. 2020. Microbial associations and spatial proximity predict North American moose (Alces alces) gastrointestinal community composition. Journal of Animal Ecology, 89 (3): 817-828. | |
| Heinritz S N, Weiss E, Eklund M, Aumiller T, Louis S, Rings A, Messner S, Camarinha‑Silva A, Seifert J, Bischoff S C, Mosenthin R. 2016. Intestinal microbiota and microbial metabolites are changed in a pig model fed a High-Fat/Low-Fiber or a Low-Fat/High-Fiber Diet. PLoS ONE, 11 (4): 0154329. | |
| Hu X L, Liu G, Shafer A B A, Wei Y T, Zhou J, Lin S, Wu H, Zhou M, Hu D F, Liu S Q. 2017. Comparative analysis of the gut microbial communities in forest and alpine musk deer using high‑throughput sequencing. Frontiers in Microbiology, 8: 572. | |
| Hu X L. 2017. The quantitative studies on the dynamics of gastrointestinal parasites and microbiota in forest musk deer and the indicative function on health. Ph.D thesis. Harbin: Northeast Forestry University. (in Chinese) | |
| Jewell K A, McCormick C A, Odt C L, Weimer P J, Suen G. 2015. Ruminal bacterial community composition in dairy cows is dynamic over the course of two lactations and correlates with feed efficiency. Applied and Environmental Microbiology, 81 (14): 4697-4710. | |
| Jiang F, Gao H M, Qin W, Song P F, Wang H J, Zhang J J, Liu D X, Wang D, Zhang T Z. 2021. Marked seasonal variation in structure and function of gut microbiota in forest and alpine musk deer. Frontiers in Microbiology, 12: 699797. | |
| La Reau A J, Meier‑Kolthoff J P, Suen G. 2016. Sequence-based analysis of the genus Ruminococcus resolves its phylogeny and reveals strong host association. Microbial Genomics, 2 (12): 1-13. | |
| La Reau A J, Suen G. 2018.The ruminococci: key symbionts of the gut ecosystem. Journal of Microbiology, 56 (3): 199-208. | |
| Lang J M, Pan C, Cantor R M, Tang W H W, Garcia‑Garcia J C, Kurtz I, Hazen S L, Bergeron N, Krauss R M, Lusis A J. 2018. Impact of individual traits, saturated fat, and protein source on the gut microbiome. mBio, 9 (6): e01604-18. | |
| Ley R E, Peterson D A, Gordon J I. 2006. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell, 124 (4): 837-848. | |
| Li Y M, Hu X L, Yang S, Zhou J T, Zhang T X, Qi L, Sun X N, Fan M Y, Xu S H, Cha M H, Zhang M S, Lin S B, Liu S Q, Hu D F. 2017. Comparative analysis of the gut microbiota composition between captive and wild forest musk deer. Frontiers in Microbiology, 8: 1705. | |
| Li Y M, Hu X L, Yang S, Zhou J T, Qi L, Sun X M, Fan M Y, Xu S H, Cha M H, Zhang M S, Lin S B, Liu S Q, Hu D F. 2018. Comparison between the fecal bacterial microbiota of healthy and diarrheic captive musk deer. Frontiers in Microbiology, 9: 300. | |
| Liu X, Zhao W, Yu D, Cheng J G, Luo Y, Wang Y, Yang Z X, Yao X P, Wu S S, Wang W Y, Yang W, Li D Q, Wu Y M. 2019. Effects of compound probiotics on the weight, immunity performance and fecal microbiota of forest musk deer. Scientific Reports, 9 (1): 19146. | |
| Lv X H, Qiao J Y, Wu X M, Su L N. 2009. A review of mainly affected on musk‑deer diseases: purulent, respiratory system and parasitic diseases. Journal of Economic Animal, 13 (2): 104-107. (in Chinese) | |
| Magoč T, Salzberg S L. 2021. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27 (21): 2957-2963. | |
| Matulova M, Nouaille R, Capek P, Péan M, Delort A M, Forano E. 2008. NMR study of cellulose and wheat straw degradation by Ruminococcus albus 20. FEBS Journal, 275 (13): 3503-3511. | |
| Nicholson J K, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-Gut microbiota metabolic interactions. Science, 336 (6086): 1262-1267. | |
| O’Toole P W, Jeffery I B. 2015. Gut microbiota and aging. Science, 350 (6265): 1214-1215. | |
| Shanahan F. 2010. Probiotics in perspective. Gastroenterology, 139 (6): 1808-1812. | |
| Sun X N, Cai R B, Jin X L, Shafer A B A, Hu X L, Yang S, Li Y M, Qi L, Liu S Q, Hu D F. 2018. Blood transcriptomics of captive forest musk deer (Moschus berezovskii) and possible associations with the immune response to abscesses. Scientific Reports, 8 (1): 599. | |
| Wan Y, Wang F L, Yuan J H, Li J, Jiang D D, Zhang J J, Li H, Wang R Y, Tang J, Huang T, Zheng J S, Sinclair A J, Mann J, Li D. 2019. Effects of dietary fat on gut microbiota and faecal metabolites, and their relationship with cardiometabolic risk factors: a 6-month randomised controlled-feeding trial. Gut, 68 (8): 1417-1429. | |
| Wang Q, Garrity G M, Tiedje J M, Cole J R. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology, 73 (16): 5261-5267. | |
| Wang Y Y, Cao P H, Wang L, Zhao Z Y, Chen Y L, Yang Y X. 2017. Bacterial community diversity associated with different levels of dietary nutrition in the rumen of sheep. Applied Microbiology and Biotechnology, 101 (9): 3717-3728. | |
| Waters J L, Ley R E. 2019. The human gut bacteria Christensenellaceae are widespread, heritable, and associated with health. BMC Biology, 17 (1): 83. | |
| Wu S R, Cui Z H, Chen X D, Zheng L X, Ren H, Wang D D, Yao J H. 2021. Diet-ruminal microbiome-host crosstalk contributes to differential effects of calf starter and alfalfa hay on rumen epithelial development and pancreatic α-amylase activity in yak calves - ScienceDirect. Journal of Dairy Science, 104 (4): 4326-4340. | |
| Yang Q S, Meng X X, Xia L, Feng Z J. 2003. Conservation status and causes of decline of musk deer (Moschus spp.) in China. Biological Conservation, 109 (3): 333-342. | |
| Yin J, Li Y Y, Han H, Chen S, Gao J, Liu G, Wu X, Deng J P, Yu Q F, Huang X G, Fang R J, Li T J, Reiter R J, Zhang D, Zhu C R, Zhu G Q, Ren W K, Yin Y L. 2018. Melatonin reprogramming of gut microbiota improves lipid dysmetabolism in high-fat diet-fed mice. Journal of Pineal Research, 65 (4): e12524. | |
| Yoo J Y, Groer M, Dutra S V O, Sarkar A, McSkimming D I. 2020. Gut microbiota and immune system interactions. Microorganisms, 8 (10): 1587. | |
| Zhang C H, Zhang M H, Wang S Y, Han R J, Cao Y F, Hua W Y, Mao Y J, Zhang X J, Pang X Y, Wei C C, Zhao G P, Chen Y, Zhao L P. 2010. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME Journal, 4 (2): 232-241. | |
| Zhang Z G, Xu D M, Wang L, Hao J J, Wang J F, Zhou X, Wang W W, Qiu Q, Huang X D, Zhou J W, Long R F, Zhao F Q, Shi P. 2016. Convergent evolution of rumen microbiomes in high‑altitude mammals. Current Biology, 26 (14): 1873-1879. | |
| Zhao X F, Zhang X Y, Chen Z S, Wang Z, Lu Y Y, Cheng D F. 2018. The divergence in bacterial components associated with Bactrocera dorsalis across developmental stages. Frontiers in Microbiology, 9: 114. | |
| Zhou C, Zhang W B, Wen Q C, Bu P, Gao J, Wang G N, Jin J Z, Song Y J, Sun X H, Zhang Y F, Jiang X, Yu H R, Peng C J, Shen Y M, Price M, Li J, Zhang X Y, Fan Z X, Yue B S. 2019. Comparative genomics reveals the genetic mechanisms of musk secretion and adaptive immunity in Chinese forest musk deer. Genome Biology and Evolution, 11 (4): 1019-1032. | |
| Zhou L Y, Xiao X H, Zhang Q, Zheng J, Li M, Yu M, Wang X J, Deng M Q, Zhai X, Li R R. 2018. Improved glucose and lipid metabolism in the early life of female offspring by maternal dietary genistein is associated with alterations in the gut microbiota. Frontiers in Endocrinology, 9: 516. | |
| 吕向辉, 乔继英, 吴晓民, 苏丽娜. 2009. 主要危害麝类的化脓性、呼吸系统疾病及寄生虫病研究. 经济动物学报, 13 (2): 104-107. | |
| 吴家炎, 王伟. 2006. 中国麝类. 北京: 中国林业出版社. | |
| 国家林业局. 2009. 中国重点陆生野生动物资源调查. 北京: 中国林业出版社. | |
| 胡晓龙. 2017. 林麝肠道寄生虫和菌群动态的量化研究及其健康指示作用. 哈尔滨: 东北林业大学博士学位论文. |
| [1] | 郑凯丹, 汪巧云, 范朋飞, 韩雪松, 肖梅, 谌利民, 董正一, 张璐. 基于微卫星的欧亚水獭个体识别和遗传多样性[J]. 兽类学报, 2024, 44(2): 146-158. |
| [2] | 赛青高娃, 王子涵, 李全邦, 王东, 连新明. 藏羚交配群成员间亲缘关系及遗传多样性分析[J]. 兽类学报, 2024, 44(2): 159-170. |
| [3] | 黄小龙, 李海波, 张旭, 程绍传, 晏玉莹, 杨伟, 蒙秉顺, 王丞, 杨杰, 冉景丞. 梵净山同域分布黔金丝猴与藏酋猴的肠道微生物结构差异[J]. 兽类学报, 2024, 44(2): 183-194. |
| [4] | 袁道欢, 程寿杰, 袁倩敏, 纪羽, 秦姣, 梁易天, 刘全生. 深圳机场小型兽类组成、多样性及时空变化[J]. 兽类学报, 2024, 44(2): 252-258. |
| [5] | 余苗杰, 徐忠鲜, 蒋雪梅, 王春花, 赵婵娟, 戚文华, 竭航. 林麝不同组织miRNA表达谱[J]. 兽类学报, 2023, 43(6): 723-733. |
| [6] | 张如梅, 张庆, 杨逍, 张发瑞, 赵定, 庞德洪, 杨孔, 官天培. 高山生态系统哺乳动物多样性时空分布格局——以大熊猫国家公园雪宝顶片区为例[J]. 兽类学报, 2023, 43(5): 533-543. |
| [7] | 平晓鸽, 朱江, 魏辅文. 从埃及到昆明-蒙特利尔——2020年后全球生物多样性框架的转变[J]. 兽类学报, 2023, 43(4): 357-363. |
| [8] | 刘玲, 肖爱刚, 赵铁建, 冯小梅, 沈素侠, 卢宪旺, 关洪武, 赵大鹏. 基于红外相机技术的野生豹猫行为谱和PAE编码系统[J]. 兽类学报, 2023, 43(3): 270-279. |
| [9] | 邹书珍, 罗亚, 程鸣, 王凡, 黎大勇, 康迪, 唐贇. 滇金丝猴肠道微生物四环素抗性基因和肠道环境特征[J]. 兽类学报, 2023, 43(3): 304-314. |
| [10] | 阮向东, 陈奕欣, 王博宇, 杨筱, 廖春林, 禹洋, 郭程. 湖南八大公山国家级自然保护区兽类和鸟类多样性及活动节律调查[J]. 兽类学报, 2023, 43(3): 342-351. |
| [11] | 张同作, 江峰, 张婧捷, 蔡振媛, 高红梅, 顾海峰, 宋鹏飞. 三江源国家公园野生动物保护与管理对策[J]. 兽类学报, 2023, 43(2): 193-205. |
| [12] | 谢博, 农秀萍, 黄国力, 黄蓉, 姚维, 林建忠, 周岐海. 广西恩城国家级自然保护区兽类和鸟类多样性红外相机监测初报[J]. 兽类学报, 2023, 43(2): 215-223. |
| [13] | 张清浩, 姚松, 徐恺, 刘统, 肖文宏, 白兵勇, 黄小群, 肖治术. 基于红外相机技术对河南内乡宝天曼国家级自然保护区鸟兽多样性的调查[J]. 兽类学报, 2023, 43(2): 206-214. |
| [14] | 田新民, 张明海. 基于粪便DNA的东北马鹿遗传多样性[J]. 兽类学报, 2023, 43(1): 41-49. |
| [15] | 郭秋艳, 韦晓, 陆媚静, 范鹏来, 周岐海. 非人灵长类肠道微生物研究进展与展望[J]. 兽类学报, 2023, 43(1): 69-81. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2