

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2022, Vol. 42 ›› Issue (4): 432-441.DOI: 10.16829/j.slxb.150637
王小琪1,2,3, 郝文静1, 张志超1, 韩晶1,4, 王儒敬2,3, 段子渊1( )
)
收稿日期:2021-11-15
接受日期:2022-05-06
出版日期:2022-07-30
发布日期:2022-07-22
通讯作者:
段子渊
作者简介:王小琪 (1989- ),女,博士研究生,主要从事肠道微生物组研究.
基金资助:
Xiaoqi WANG1,2,3, Wenjing HAO1, Zhichao ZHANG1, Jing HAN1,4, Rujing WANG2,3, Ziyuan DUAN1( )
)
Received:2021-11-15
Accepted:2022-05-06
Online:2022-07-30
Published:2022-07-22
Contact:
Ziyuan DUAN
摘要:
为比较青藏高原柴达木马亚成体腹泻与健康个体粪便微生物群落多样性和结构组成的差异, 我们利用16S rRNA测序技术对采集的腹泻 (n = 3) 和健康 (n = 13) 个体粪便样本细菌的组成与分布进行分析比较,并利用实时荧光定量PCR测定相关菌属的含量。结果显示,无论健康还是腹泻,厚壁菌门、拟杆菌门、疣微菌门、变形杆菌门和螺旋体门是柴达木马亚成体粪便中的优势菌门。相比健康组,腹泻组粪便微生物的Alpha多样性显著下降 (P < 0.05),厚壁菌门相对丰度下降而变形杆菌门的相对丰度显著增加 (P < 0.05),推断这两个门中的梭菌属、普雷沃菌属、纤杆菌属等丰度的失衡可能是导致柴达木马腹泻的原因之一。此外,通过机器学习的随机森林算法筛选出12个对健康和腹泻柴达木马亚成体粪便微生物差异具有较大影响的特征菌属,包括甲烷短杆菌属、纤杆菌属、Paludibacter、肉食杆菌属和迷踪菌属等。研究揭示了健康和腹泻柴达木马亚成体粪便微生物组的变化,为进一步研究青藏高原地区家畜腹泻提供一定的数据支持。
中图分类号:
王小琪, 郝文静, 张志超, 韩晶, 王儒敬, 段子渊. 腹泻柴达木马亚成体粪便微生物多样性分析及其生物标记物的筛选[J]. 兽类学报, 2022, 42(4): 432-441.
Xiaoqi WANG, Wenjing HAO, Zhichao ZHANG, Jing HAN, Rujing WANG, Ziyuan DUAN. Fecal microbiota diversity analysis of the diarrheal sub-adult Qaidam horses and biomarkers screening[J]. ACTA THERIOLOGICA SINICA, 2022, 42(4): 432-441.
| 属Genera | 引物Primers (5'-3') | 参考文献References |
|---|---|---|
拟杆菌属 Bacteroides | F: CTGAACCAGCCAAGTAGCG | |
| R: CCGCAAACTTTCACAACTGACTTA | ||
普雷沃菌属 Prevotella | F: GGTTCTGAGAGGAAGGTCCCC | |
| R: TCCTGCACGCTACTTGGCTG | ||
甲烷短杆菌属 Methanobrevibacter | F: CCTCCGCAATGTGAGAAATCGC | |
| R: TCWCCAGCAATTCCCACCAGTT | ||
双歧杆菌属 Bifidobacterium | F: CGCGTCYGGTGTGAAAG | |
| R: CCCCACATCCAGCATCCA |
表1 柴达木马粪便细菌16S rRNA 基因PCR扩增引物
Table 1 PCR amplification primers of bacterial 16S rRNA gene in Qaidam horses’ feces
| 属Genera | 引物Primers (5'-3') | 参考文献References |
|---|---|---|
拟杆菌属 Bacteroides | F: CTGAACCAGCCAAGTAGCG | |
| R: CCGCAAACTTTCACAACTGACTTA | ||
普雷沃菌属 Prevotella | F: GGTTCTGAGAGGAAGGTCCCC | |
| R: TCCTGCACGCTACTTGGCTG | ||
甲烷短杆菌属 Methanobrevibacter | F: CCTCCGCAATGTGAGAAATCGC | |
| R: TCWCCAGCAATTCCCACCAGTT | ||
双歧杆菌属 Bifidobacterium | F: CGCGTCYGGTGTGAAAG | |
| R: CCCCACATCCAGCATCCA |
组别 Groups | 有效序列片段数 No. of filtered reads | 操作分类单元数 OTUs | 分类单元(属) Taxa (genus) | 样本覆盖度 Coverage |
|---|---|---|---|---|
| 腹泻组 Diarrhea | 117 711 | 4 501 | 149 | 0.975 |
| 健康组 Naive control | 457 643 | 6 030 | 164 | 0.972 |
表2 柴达木马粪便细菌16S rRNA测序数据
Table 2 16S rRNA sequencing data of feces from Qaidam horses
组别 Groups | 有效序列片段数 No. of filtered reads | 操作分类单元数 OTUs | 分类单元(属) Taxa (genus) | 样本覆盖度 Coverage |
|---|---|---|---|---|
| 腹泻组 Diarrhea | 117 711 | 4 501 | 149 | 0.975 |
| 健康组 Naive control | 457 643 | 6 030 | 164 | 0.972 |
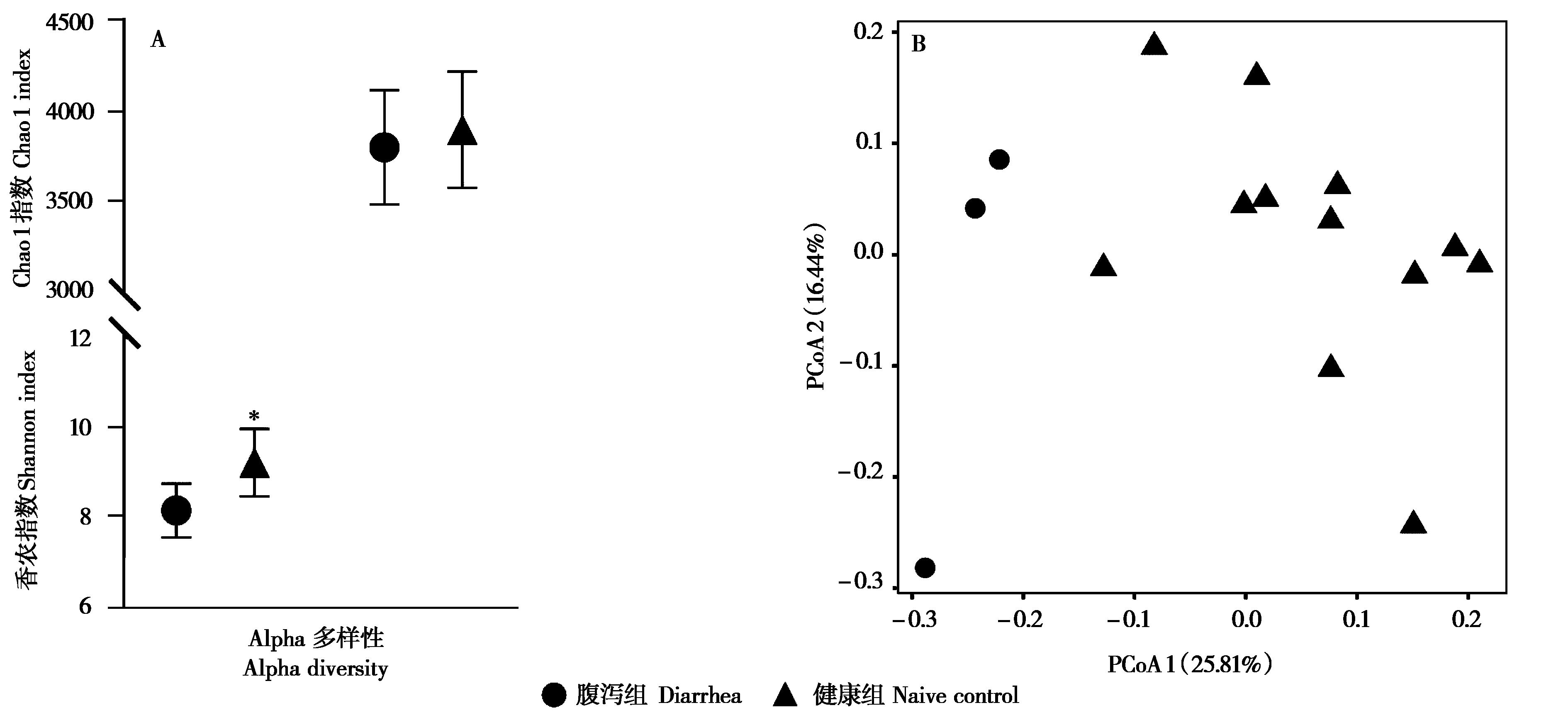
图1 柴达木马粪便微生物物种多样性.A:粪便微生物群落Alpha多样性 (均值 ± 标准差);B:基于非加权Unifrac矩阵的主坐标分析图.* P < 0.05
Fig. 1 The fecal microbial diversity of Qaidam horses. A: The Alpha diversity of fecal bacteria (mean ± SD); B: PCoA plot based on Unweighted Unifrac matrix.* P < 0.05
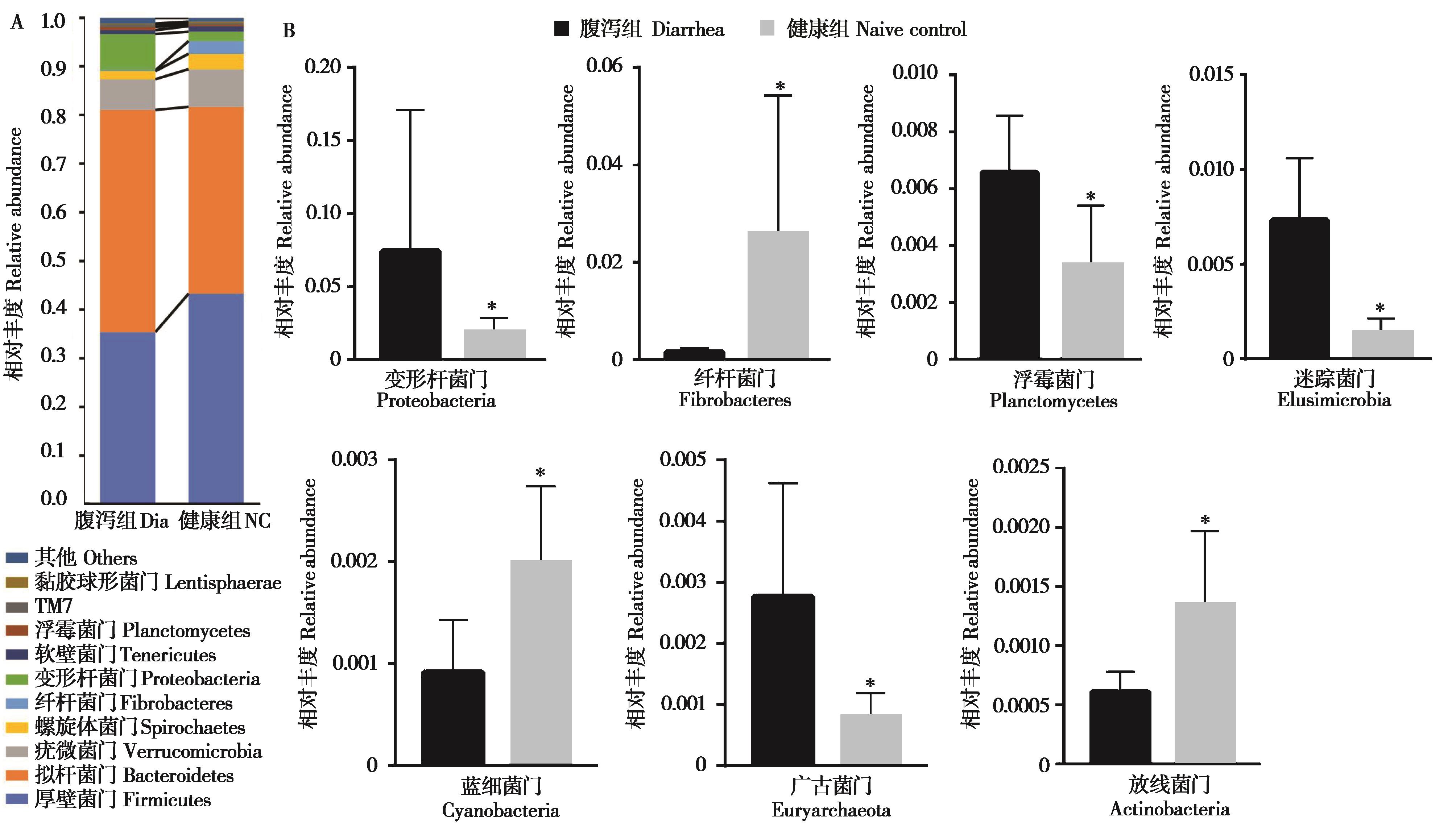
图2 腹泻组和健康组柴达木马粪便微生物门水平的相对丰度比较. A:粪便细菌相对丰度组成 (门水平);B:两组间有显著性差异的菌门. * P < 0.05
Fig. 2 Comparison of relative abundance of fecal microbial in diarrhea (Dia) group and naive control (NC) group of Qaidam horses at phylum level. A: The histograms of microbiota abundances at phylum level; B: The phyla with significant difference between two groups. * P < 0.05
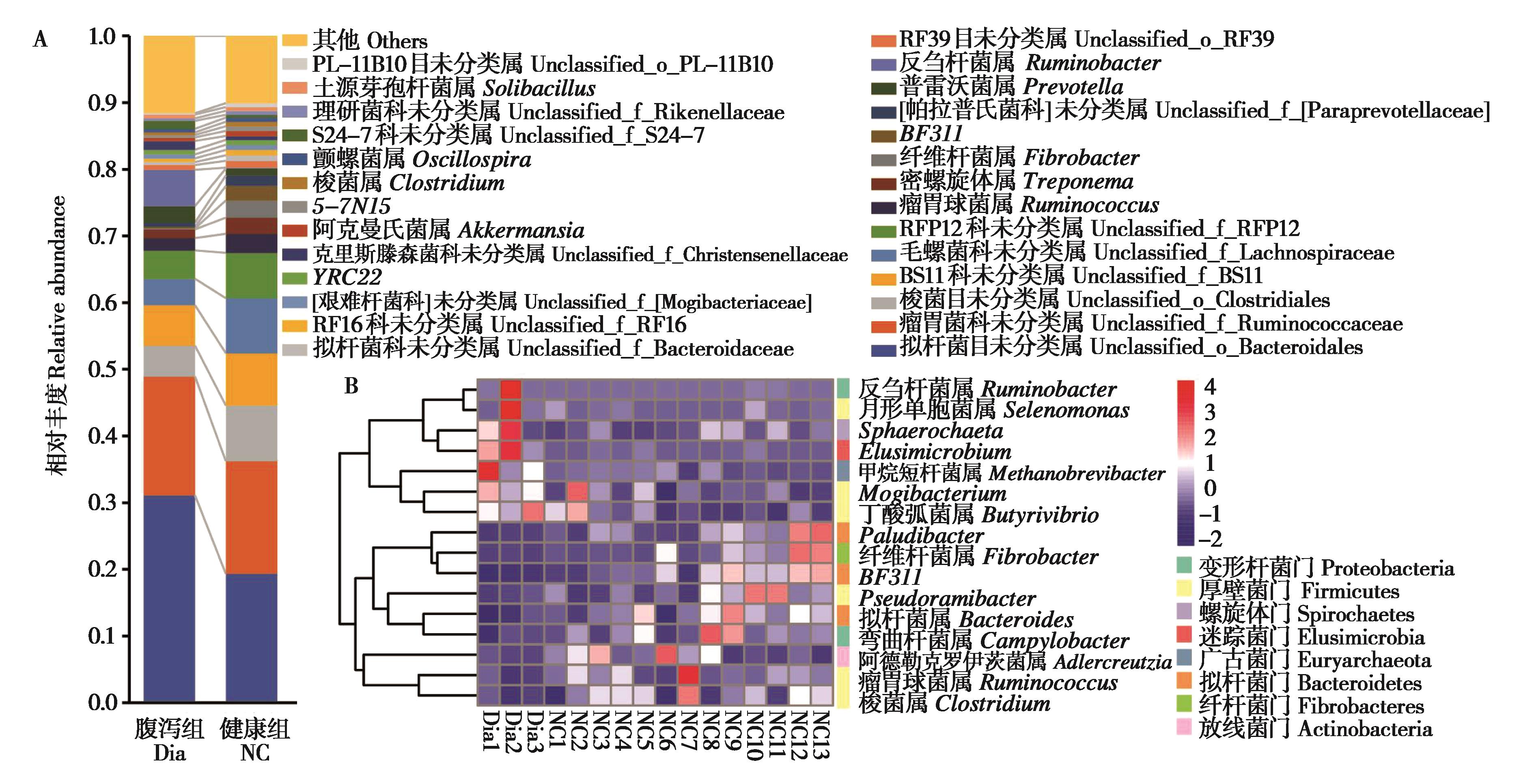
图3 腹泻组和健康组柴达木马粪便微生物属水平的相对丰度比较. A:粪便细菌相对丰度组成 (属水平);B:热图展示的两组间有显著性差异的菌属
Fig. 3 Comparison of relative abundance of fecal microbial in diarrhea (Dia) group and naive control (NC) group of Qaidam horses at genus level. A: The histograms of microbiota abundance at genus level; B: Clustering heatmap of genera with significant difference between two groups

图4 腹泻组和健康组柴达木马粪便主要菌属16S rRNA基因拷贝数差异分析
Fig. 4 Compared average copy number of 16S rRNA gene from the main fecal bacteria between diarrhea group and naive control group of Qaidam horses
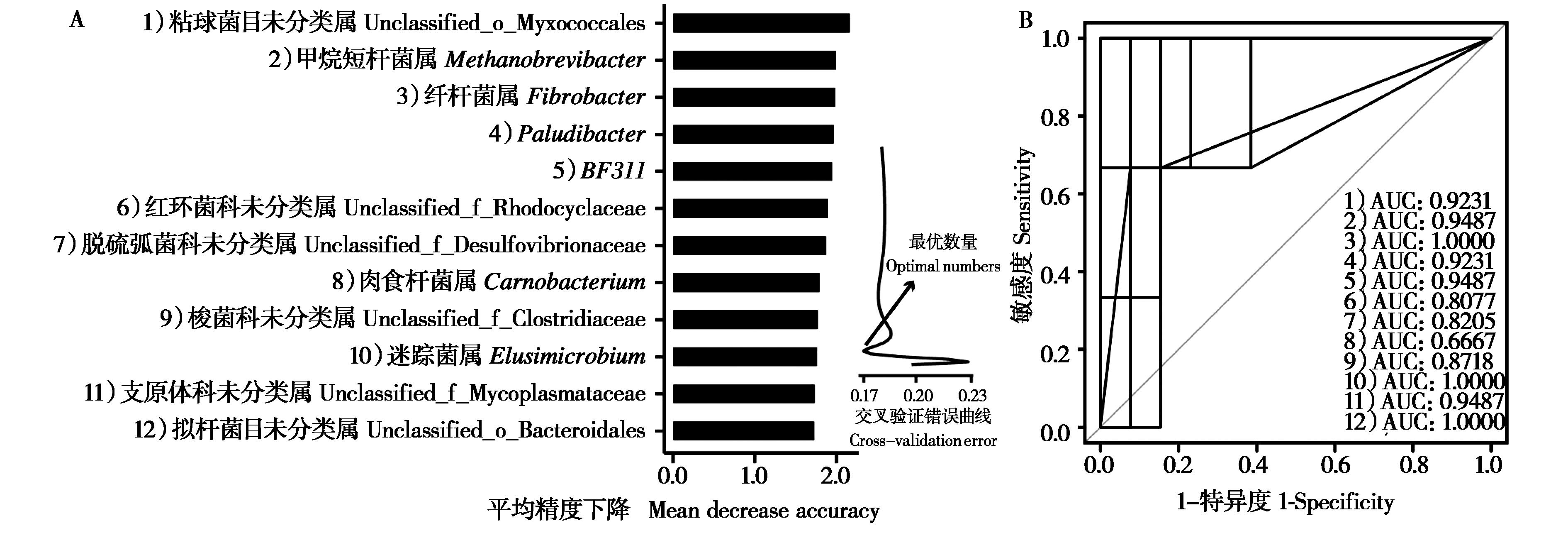
图5 基于随机森林的健康和腹泻柴达木马粪便微生物差异分析 (A) 及ROC曲线分析 (B)
Fig. 5 Random Forest bar chart of variable importance on genus level (biomarkers) (A) and ROC curve analysis (B)
| Al Jassim R A, Andrews F M. 2009. The bacterial community of the horse gastrointestinal tract and its relation to fermentative acidosis, laminitis, colic, and stomach ulcers. Veterinary Clinics of North America: Equine Practice, 25 (2): 199-215. | |
| Chen S B, Ouyang Z Y, Xu W H, Xiao Y. 2010. A review of beta diversity studies. Biodiversity Science, 18 (4): 323-335. (in Chinese) | |
| Chung W, Walker A W, Bosscher D, Garcia‑Campayo V, Wagner J, Parkhill J, Duncan S H, Flint H J. 2020. Relative abundance of the prevotella genus within the human gut microbiota of elderly volunteers determines the inter‑individual responses to dietary supplementation with wheat bran arabinoxylan-oligosaccharides. BMC Microbiology, 20 (1): 283. | |
| Costa M C, Weese J S. 2018. Understanding the intestinal microbiome in health and disease. Veterinary Clinics of North America: Equine Practice, 34 (1): 1-12. | |
| Costa M C, Arroyo L G, Allen‑Vercoe E, Stämpfli H R, Kim P T, Sturgeon A, Weese J S. 2012. Comparison of the fecal microbiota of healthy horses and horses with colitis by high throughput sequencing of the V3-V5 region of the 16S rRNA gene. PLoS ONE, 7 (7): e41484. | |
| Dong Z G, Liu Y B, Pan H W, Wang H C, Wang X, Xu X F, Xiao K, Liu M, Xu Z Y, Li L B, Zhang Y. 2020. The effects of high‑salt gastric intake on the composition of the intestinal microbiota in Wistar rats.Medical Science Monitor: International Medical Journal of Experimental and Clinical Research, 26: e922160. | |
| Drancourt M, Prebet T, Aghnatios R, Edouard S, Cayrou C, Henry M, Blaise D, Raoult D. 2014. Planctomycetes DNA in febrile aplastic patients with leukemia, rash, diarrhea, and micronodular pneumonia. Journal of Clinical Microbiology, 52 (9): 3453-3455. | |
| Fassarella M, Blaak E E, Penders J, Nauta A, Smidt H, Zoetendal E G. 2021. Gut microbiome stability and resilience: elucidating the response to perturbations in order to modulate gut health. Gut, 70 (3): 595-605. | |
| Garber A, Hastie P, Murray J A. 2020. Factors influencing equine gut microbiota: current knowledge. Journal of Equine Veterinary Science, 88: 102943. | |
| Biomarkers Definitions Working Group. 2001. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clinical Pharmacology and Therapeutics, 69 (3): 89-95. | |
| Herlemann D P, Labrenz M, Jürgens K, Bertilsson S, Waniek J J, Andersson A F. 2011. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. The ISME Journal, 5 (10): 1571-1579. | |
| Jiang M. 2014. Analysis of difference in intestinal flora of Uygur and Han ethnic Chinese patients with ulcerative colitis. Master thesis. Urumqi: Xinjiang Medical University. (in Chinese) | |
| Kameoka S, Motooka D, Watanabe S, Kubo R, Jung N, Midorikawa Y, Shinozaki N O, Sawai Y, Takeda A K, Nakamura S. 2021. Benchmark of 16S rRNA gene amplicon sequencing using Japanese gut microbiome data from the V1-V2 and V3-V4 primer sets. BMC Genomics, 22 (1): 527. | |
| Kovatcheva‑Datchary P, Nilsson A, Akrami R, Lee Y S, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. 2015.Dietary fiber‑induced improvement in glucose metabolism is associated with increased abundance of Prevotella . Cell Metabolism, 22 (6): 971-982. | |
| Li D L, Sun Y, Xu F R. 2021. Differences in faeces microbiome composition and characteristics between two populations of Ptyas dhumnades . Chinese Journal of Zoology, 56 (5): 696-706. (in Chinese) | |
| Li L. 2020. Effect of different levels of tannin in diets on ruminal bacteria number and methanogens diversity in sheep. Master thesis. Hohhot: Inner Mongolia Agricultural University. (in Chinese) | |
| Liu B D, Lin W F, Chen S J, Xiang T, Yang Y F, Yin Y L, Xu G H, Liu Z H, Liu L, Pan J Y, Xie L W. 2019. Gut microbiota as an objective measurement for auxiliary diagnosis of insomnia disorder. Frontiers in Microbiology, 10: 1770. | |
| Mei L J, Zhou J L, Su Y M, Mao K H, Wu J, Zhu C C, He L, Cui Y. 2021. Gut microbiota composition and functional prediction in diarrhea-predominant irritable bowel syndrome. BMC Gastroenterol, 21 (1): 105. | |
| Metzler B U, Mosenthin R. 2008. A review of interactions between dietary fiber and the gastrointestinal microbiota and their consequences on intestinal phosphorus metabolism in growing pigs. Asian Australasian Journal of Animal Sciences, 21: 603-615. | |
| Moreno‑Indias I, Sánchez‑Alcoholado L, Pérez‑Martínez P, Andrés‐Lacueva C, Cardona F, Tinahones F, Queipo‑Ortuño M I. 2016. Red wine polyphenols modulate fecal microbiota and reduce markers of the metabolic syndrome in obese patients. Food Funct, 7 (4): 1775-1787. | |
| Netherwood T, Wood J L, Townsend H G, Mumford J A, Chanter N. 1996. Foal diarrhoea between 1991 and 1994 in the United Kingdom associated with Clostridium perfringens, rotavirus, Strongyloides westeri and Cryptosporidium spp. Epidemiology and Infection, 117 (2): 375-383. | |
| Oliver‑Espinosa O. 2018. Foal diarrhea: established and postulated causes, prevention, diagnostics, and treatments. Veterinary Clinics of North America: Equine Practice, 34 (1): 55-68. | |
| Pandit R J, Hinsu A T, Patel S H, Jakhesara S J, Koringa P G, Bruno F, Psifidi A, Shah S V, Joshi C G. 2018. Microbiota composition, gene pool and its expression in Gir cattle (Bos indicus) rumen under different forage diets using metagenomic and metatranscriptomic approaches. Systematic and Applied Microbiology, 41 (4): 374-385. | |
| Rodriguez C, Taminiau B, Brévers B, Avesani V, Van Broeck J, Leroux A, Gallot M, Bruwier A, Amory H, Delmée M, Daube G. 2015. Faecal microbiota characterisation of horses using 16 rDNA barcoded pyrosequencing, and carriage rate of clostridium difficile at hospital admission. BMC Microbiology, 15: 181. | |
| Schoster A, Staempfli H R, Guardabassi L G, Jalali M, Weese J S. 2017. Comparison of the fecal bacterial microbiota of healthy and diarrheic foals at two and four weeks of life. BMC Veterinary Research, 13 (1): 144. | |
| Shaw S D, Stämpfli H. 2018. Diagnosis and treatment of undifferentiated and infectious acute diarrhea in the adult horse. Veterinary Clinics of North America: Equine Practice, 34 (1): 39-53. | |
| Shin N R, Whon T W, Bae J W. 2015. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends in Biotechnology, 33 (9): 496-503. | |
| Stamps B W, Lyon W J, Irvin A P, Kelley‑Loughnane N, Goodson M S. 2020. A pilot study of the effect of deployment on the gut microbiome and Traveler’s Diarrhea susceptibility. Frontiers Cellular and Infection Microbiology, 10: 589297. | |
| Stern E K, Brenner D M. 2018. Gut microbiota‑based therapies for irritable bowel syndrome. Clinical and Translational Gastroenterology, 9 (2): e134. | |
| Wang B, Deng B, Yong F, Zhou H X, Qu C P, Zhou Z Y. 2020. Comparison of the fecal microbiomes of healthy and diarrheic captive wild boar. Microbial Pathogensis, 147: 104377. | |
| Wang X Q, Zhang Z C, Wang X P, Bao Q, Wang R J, Duan Z Y. 2021. The impact of host genotype, intestinal sites and probiotics supplementation on the gut microbiota composition and diversity in sheep. Biology (Basel), 10 (8): 769. | |
| Ward T, Larson J, Meulemans J, Hillmann B, Lynch J, Sidiropoulos D, Spear J R, Caporaso G, Blekhman R, Knight R, Fink R, Knights D. 2017. BugBase predicts organism-level microbiome phenotypes. bioRxiv: 133462. | |
| Xi L, Song Y M, Han J C, Qin X X. 2021. Microbiome analysis reveals the significant changes in gut microbiota of diarrheic Baer’s Pochards (Aythya baeri). Microbial Pathogenesis, 157: 105015. | |
| Xie H S, Huan L, Merritt A M, Ott E A. 1997. Equine chronic diarrhea: traditional chinese veterinary medicine review. Journal of Equine Veterinary Science, 17 (12): 667-674. | |
| Xia Y, He X W, Wen X H. 2018. Mathematic methods for the evaluation of microbial diversity and their applications in the research on wastewater treatment systems. Microbiology China, 45 (8): 1778-1786. (in Chinese) | |
| Yang L N, Bian G R, Zhu W Y. 2014. Interactions between the monogastric animal gut microbiota and the intestinal immune function- A review. Acta Microbiologica Sinica, 54 (5): 480-486. (in Chinese) | |
| Zeng Q, Li D F, He Y, Li Y H, Yang Z Y, Zhao X L, Liu Y H, Wang Y, Sun J, Feng X, Wang F, Chen J X, Zheng Y J, Yang Y Y, Sun X L, Xu X M, Wang D X, Kenney T, Jiang Y Q, Gu H, Li Y L, Zhou K, Li S C, Dai W K. 2019. Discrepant gut microbiota markers for the classification of obesity‑related metabolic abnormalities. Scientific Reports, 9 (1): 13424. | |
| Zhai Q X, Tian F W, Wang G, Chen W. 2013. Progress in research on the role of intestinal microbiota in human health. Food Science, 34 (15): 337-341. (in Chinese) | |
| Zhu H, Zeng D, Wang Q, Wang N, Zeng B, Niu L L, Ni X Q. 2018. Diarrhea‑associated intestinal microbiota in captive Sichuan golden snub‑nosed monkeys (Rhinopithecus roxellana). Microbes and Environments, 33 (3): 249-256. | |
| DB 63/T962-2011. 2011. 柴达木马. 青海省: 青海省质量技术监督局. | |
| 陈圣宾, 欧阳志云, 徐卫华, 肖燚. 2010. Beta多样性研究进展. 生物多样性, 18 (4): 323-335. | |
| 翟齐啸, 田丰伟, 王刚, 陈卫. 2013. 肠道微生物与人体健康的研究进展. 食品科学, 34 (15): 337-341. | |
| 蒋曼. 2014. 新疆维吾尔族与汉族溃疡性结肠炎患者肠道菌群差异性比较. 乌鲁木齐: 新疆医科大学硕士学位论文. | |
| 李栋梁, 孙越, 徐发荣. 2021. 乌梢蛇粪便微生物群组成和特征的种群间差异. 动物学杂志, 56 (5): 696-706. | |
| 丽丽. 2020. 饲粮中不同水平单宁对绵羊瘤胃细菌菌群数量及产甲烷菌多样性的影响. 呼和浩特: 内蒙古农业大学硕士学位论文. | |
| 夏瑜, 何绪文, 文湘华. 2018. 微生物群落多样性数学表征方法及其在污水处理系统研究中的应用. 微生物学通报, 45 (8): 1778-1786. | |
| 杨利娜, 边高瑞, 朱伟云. 2014. 单胃动物肠道微生物菌群与肠道免疫功能的相互作用. 微生物学报, 54 (5): 480-486. |
| [1] | 赵星, 马锐, 吴蔚, 李明喜, 陈超, 周延山, 洪明生, 齐敦武. 不同生活环境下小熊猫皮质醇和肠道微生物的变化规律[J]. 兽类学报, 2024, 44(4): 427-435. |
| [2] | 刘传发 张良志 付海波 李文靖 张贺 李吉叶 皮立 张堰铭. 野牦牛和家牦牛粪便菌群与短链脂肪酸关系的研究[J]. 兽类学报, 2019, 39(1): 1-7. |
| [3] | 徐金会 耿晓翠 陈蕾 薛慧良 徐来祥. 生长抑素基因在不同性别和年龄黑线仓鼠哈氏腺的差异表达[J]. 兽类学报, 2016, 36(4): 429-437. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2