

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2023, Vol. 43 ›› Issue (2): 182-192.DOI: 10.16829/j.slxb.150724
收稿日期:2022-08-13
接受日期:2022-11-07
出版日期:2023-03-30
发布日期:2023-03-23
通讯作者:
韦力
作者简介:邵伟伟 (1981- ),女,硕士,主要从事动物学研究.
基金资助:
Weiwei SHAO, Fen QIAO, Wei CAI, Zhihua LIN, Li WEI( )
)
Received:2022-08-13
Accepted:2022-11-07
Online:2023-03-30
Published:2023-03-23
Contact:
Li WEI
摘要:
脊椎动物基因组含有丰富的微卫星信息。本研究对翼手目动物中的大蹄蝠全基因组及其基因的微卫星分布特征进行分析,并对含有微卫星编码序列的基因进行注释分析。结果表明,大蹄蝠全基因组大小为2.24 Gb,共含有497 883个微卫星,其中,数量和比例最多的是单碱基和二碱基重复类型,分别有173 953个 (34.94%) 和222 591个 (44.71%),相对丰度分别为77.78 loci/Mb和99.52 loci/Mb。微卫星数量从单碱基重复到六碱基重复单元最多的类型分别为 (A)n、(AC)n、(TAT)n、(TTTA)n、(AACAA)n和 (TATCTA)n,比例分别为95.14%、55.25%、38.41%、22.17%、48.68%和20.30%。不同基因区和基因间区的数量及丰度不同,其中基因间区的微卫星数量及其丰度最大,分别为322 666 个和2 541.57 loci/Mb,编码区的微卫星数量及其丰度最小,分别为1 461个和461.98 loci/Mb。基因间区和全基因组的微卫星的分布特征相似。编码区最多的微卫星类型为三碱基重复单元,外显子最多的微卫星类型为单碱基、二碱基和三碱基重复单元。在微卫星丰度分布的位置特征分析中,基因上游500 bp、外显子、内含子和基因下游500 bp各个区域微卫星丰度分别为16 400.94 loci/Mb、972.12 loci/Mb、2 180.66 loci/Mb和3 899.89 loci/Mb。大蹄蝠基因中含有微卫星的编码序列 (Coding sequence, CDS) 1 461条,被注释到的基因有1 226个。GO注释到63个主要功能基因中,并分配到26 439个GO条目。KEGG富集最显著的是信号传导通路,含有146个基因。本研究结果不仅为大蹄蝠高质量微卫星的筛选提供参考,还将进一步为翼手目其他物种的全基因组微卫星分布特征分析及其微卫星在全基因组中的生物学功能研究提供参考。
中图分类号:
邵伟伟, 乔芬, 蔡玮, 林植华, 韦力. 大蹄蝠全基因组微卫星分布特征分析[J]. 兽类学报, 2023, 43(2): 182-192.
Weiwei SHAO, Fen QIAO, Wei CAI, Zhihua LIN, Li WEI. Characteristics of microsatellite distributions in genomes of Hipposideros armiger (Chiroptera)[J]. ACTA THERIOLOGICA SINICA, 2023, 43(2): 182-192.
微卫星类型 Type of SSR | 微卫星数量 Number of SSRs | 长度 Length (bp) | 丰度Abundance (loci/Mb) | 比例 Percentage (%) |
|---|---|---|---|---|
| 单碱基 Mononucleotide | 173 953 | 2 451 423 | 77.78 | 34.94 |
| 二碱基 Dinucleotide | 222 591 | 6 114 126 | 99.52 | 44.71 |
| 三碱基 Trinucleotide | 37 566 | 800 058 | 16.8 | 7.55 |
| 四碱基 Tetranucleotide | 47 250 | 1 517 188 | 21.13 | 9.49 |
| 五碱基 Pentanucleotide | 13 134 | 310 285 | 5.87 | 2.64 |
| 六碱基 Hexanucleotide | 3 389 | 98 148 | 1.52 | 0.68 |
| SSR总数 Total of SSRs | 497 883 | 11 291 228 | 222.61 | 100 |
| 总基因组大小 Whole genome length (bp) | 2 236 581 172 | |||
| 微卫星含量 SSR content of genome (%) | 0.50 | |||
表1 大蹄蝠微卫星在全基因组中的分布情况
Table 1 Distribution of microsatellites in the genomes of Hipposideros armiger
微卫星类型 Type of SSR | 微卫星数量 Number of SSRs | 长度 Length (bp) | 丰度Abundance (loci/Mb) | 比例 Percentage (%) |
|---|---|---|---|---|
| 单碱基 Mononucleotide | 173 953 | 2 451 423 | 77.78 | 34.94 |
| 二碱基 Dinucleotide | 222 591 | 6 114 126 | 99.52 | 44.71 |
| 三碱基 Trinucleotide | 37 566 | 800 058 | 16.8 | 7.55 |
| 四碱基 Tetranucleotide | 47 250 | 1 517 188 | 21.13 | 9.49 |
| 五碱基 Pentanucleotide | 13 134 | 310 285 | 5.87 | 2.64 |
| 六碱基 Hexanucleotide | 3 389 | 98 148 | 1.52 | 0.68 |
| SSR总数 Total of SSRs | 497 883 | 11 291 228 | 222.61 | 100 |
| 总基因组大小 Whole genome length (bp) | 2 236 581 172 | |||
| 微卫星含量 SSR content of genome (%) | 0.50 | |||
模体单元 Motif length | 重复单元 Repeat unit | 微卫星数量 Microsatellites | 比例 Percentage (%) |
|---|---|---|---|
单碱基重复 Mononucleotide repeat | A | 165 494 | 95.14 |
| G | 8 459 | 4.86 | |
二碱基重复 Dinucleotide repeat | AC | 122 977 | 55.25 |
| CT | 42 659 | 19.16 | |
| GC | 1 460 | 0.66 | |
| TA | 55 495 | 24.93 | |
三碱基重复 Trinucleotide repeat | TAT | 14 429 | 38.41 |
| CAA | 7 164 | 19.07 | |
| ACC | 3 653 | 9.72 | |
| CAT | 3 408 | 9.07 | |
四碱基重复 Tetranucleotide repeat | TTTA | 10 475 | 22.17 |
| AAAC | 8 049 | 17.03 | |
| ATAG | 7 078 | 14.98 | |
| CATT | 4 959 | 10.50 | |
五碱基重复 Pentanucleotide repeat | AACAA | 6 393 | 48.68 |
| TTATT | 2 544 | 19.37 | |
| TTTCT | 795 | 6.05 | |
| TATTA | 354 | 2.70 | |
六碱基重复 Hexanucleotide repeat | TATCTA | 688 | 20.30 |
| AAACAA | 510 | 15.05 | |
| GAGAGG | 284 | 8.38 | |
| AATAAA | 133 | 3.92 |
表2 大蹄蝠最多微卫星重复单元的比较
Table 2 The most frequent microsatellite motifs found in genomes of Hipposideros armiger
模体单元 Motif length | 重复单元 Repeat unit | 微卫星数量 Microsatellites | 比例 Percentage (%) |
|---|---|---|---|
单碱基重复 Mononucleotide repeat | A | 165 494 | 95.14 |
| G | 8 459 | 4.86 | |
二碱基重复 Dinucleotide repeat | AC | 122 977 | 55.25 |
| CT | 42 659 | 19.16 | |
| GC | 1 460 | 0.66 | |
| TA | 55 495 | 24.93 | |
三碱基重复 Trinucleotide repeat | TAT | 14 429 | 38.41 |
| CAA | 7 164 | 19.07 | |
| ACC | 3 653 | 9.72 | |
| CAT | 3 408 | 9.07 | |
四碱基重复 Tetranucleotide repeat | TTTA | 10 475 | 22.17 |
| AAAC | 8 049 | 17.03 | |
| ATAG | 7 078 | 14.98 | |
| CATT | 4 959 | 10.50 | |
五碱基重复 Pentanucleotide repeat | AACAA | 6 393 | 48.68 |
| TTATT | 2 544 | 19.37 | |
| TTTCT | 795 | 6.05 | |
| TATTA | 354 | 2.70 | |
六碱基重复 Hexanucleotide repeat | TATCTA | 688 | 20.30 |
| AAACAA | 510 | 15.05 | |
| GAGAGG | 284 | 8.38 | |
| AATAAA | 133 | 3.92 |
指标 Index | 基因区Genetic region | 基因间区 Intergenic region | |||
|---|---|---|---|---|---|
编码区 CDS | 非翻译区 Untranslated | 外显子 Exon | 内含子 Intron | ||
数量 Number | 1 461 | 5 081 | 7 416 | 199 340 | 322 666 |
丰度 Abundance(loci/Mb) | 461.98 | 1 647.13 | 1 188.18 | 2 204.77 | 2 541.57 |
表3 大蹄蝠微卫星在不同基因区和基因间区的数量及丰度比较
Table 3 The number and abundance of microsatellites in different genomic regions of Hipposideros armiger
指标 Index | 基因区Genetic region | 基因间区 Intergenic region | |||
|---|---|---|---|---|---|
编码区 CDS | 非翻译区 Untranslated | 外显子 Exon | 内含子 Intron | ||
数量 Number | 1 461 | 5 081 | 7 416 | 199 340 | 322 666 |
丰度 Abundance(loci/Mb) | 461.98 | 1 647.13 | 1 188.18 | 2 204.77 | 2 541.57 |
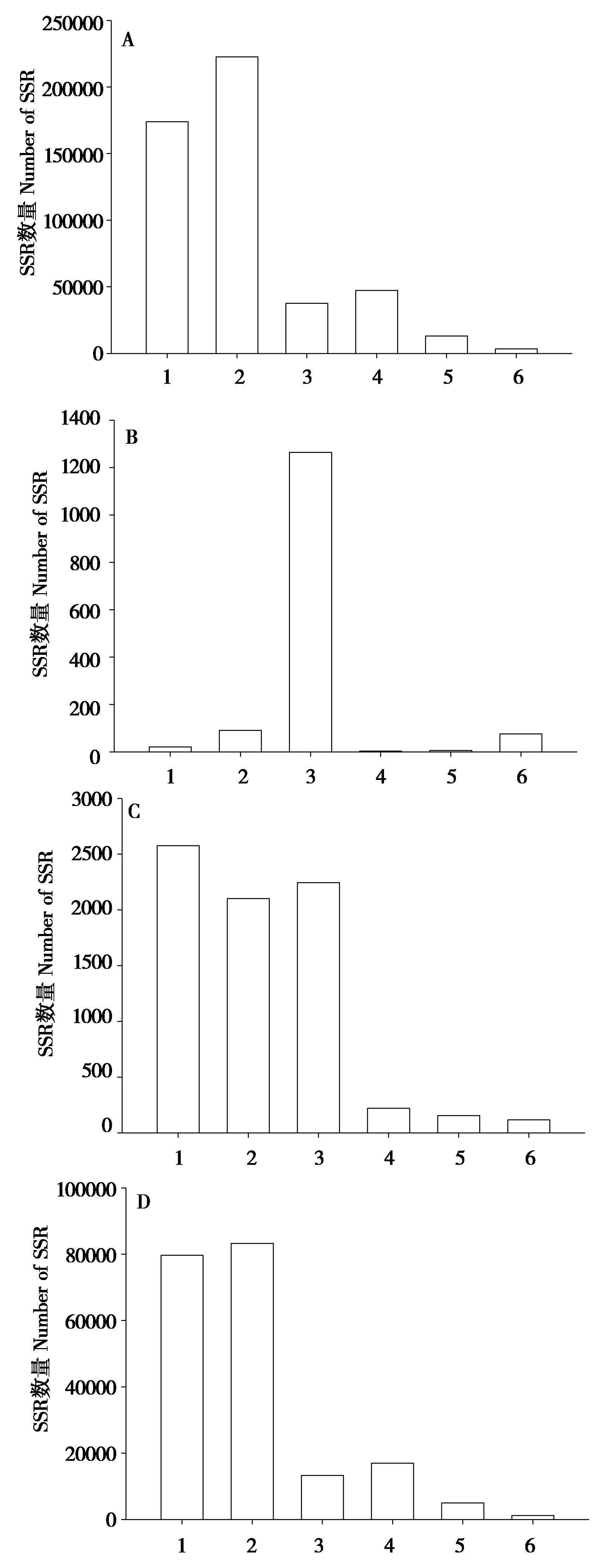
图2 大蹄蝠不同基因组区微卫星类型的分布比较. A:全基因组;B:编码区;C:外显子;D:基因间区. 1 ~ 6分别表示单碱基,二碱基,三碱基,四碱基,五碱基,六碱基
Fig. 2 The distribution of microsatellite types in different genomic regions.Genome (A), CDS (B), Exon (C) and Intergenetic (D) of Hipposideros armiger. 1 - 6 indicated mono-, di-, tri-, tetra-, penta-, hexa-nucleotide respectively
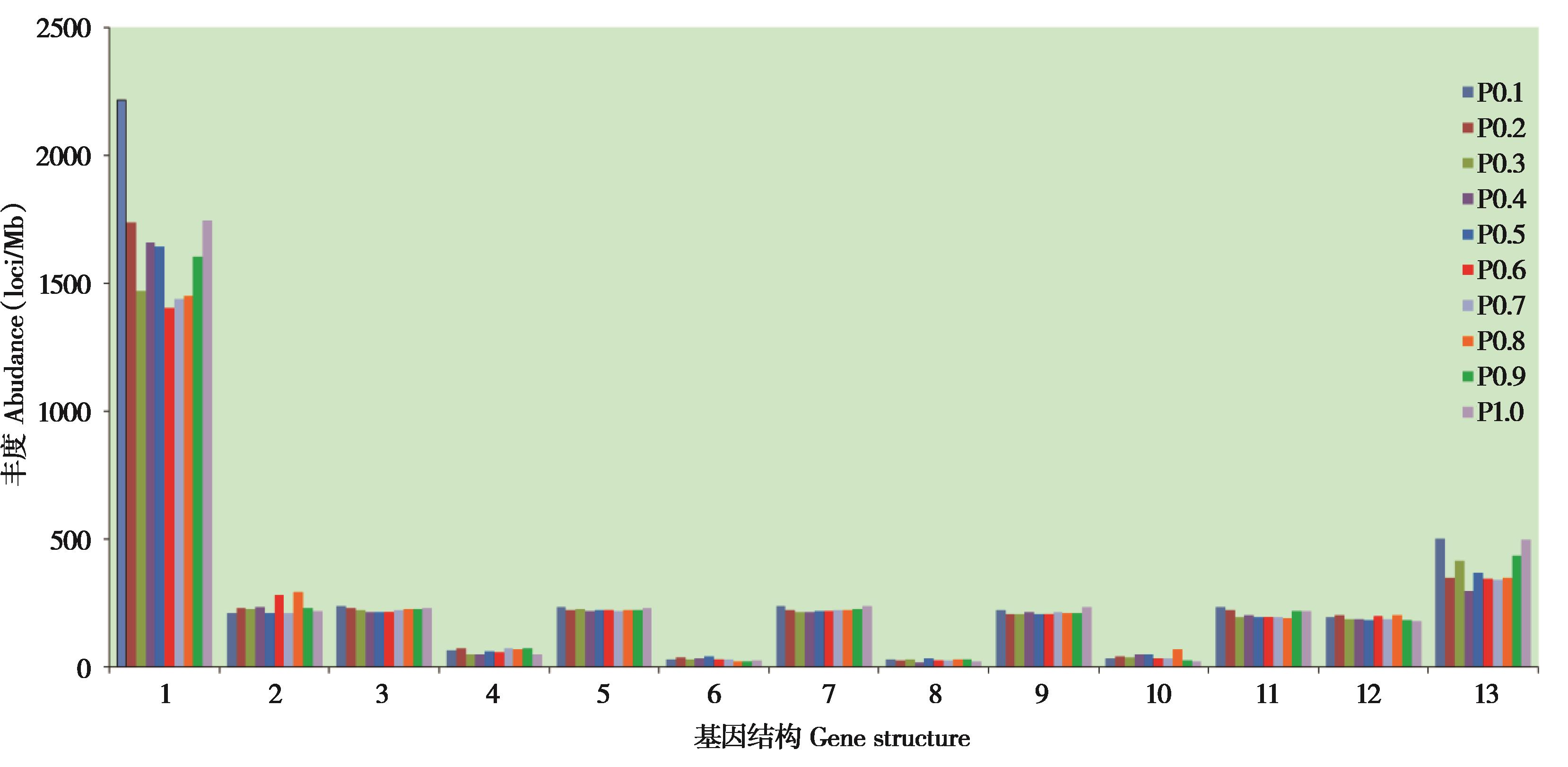
图3 大蹄蝠基因区及其上、下游微卫星丰度的比较.代码1 ~ 13分别表示基因上游500 bp、第一外显子、第一内含子、第二外显子、第二内含子、中间左外显子、中间内含子、中间右外显子、倒数第二内含子、倒数第二外显子、倒数第一内含子、倒数第一外显子、基因下游500 bp
Fig. 3 The microsatellite abundance in gene regions and their upstream and downstream regions of Hipposideros armiger. Number 1 - 13 indicated upstream 500 bp, exon 1, intron1, exon 2, intron 2, mid left exon, mid intron, mid right exon,intron reverse 2, exon reverse 2, intron reverse 1, exon reverse 1, downstream 500 bp, respectively
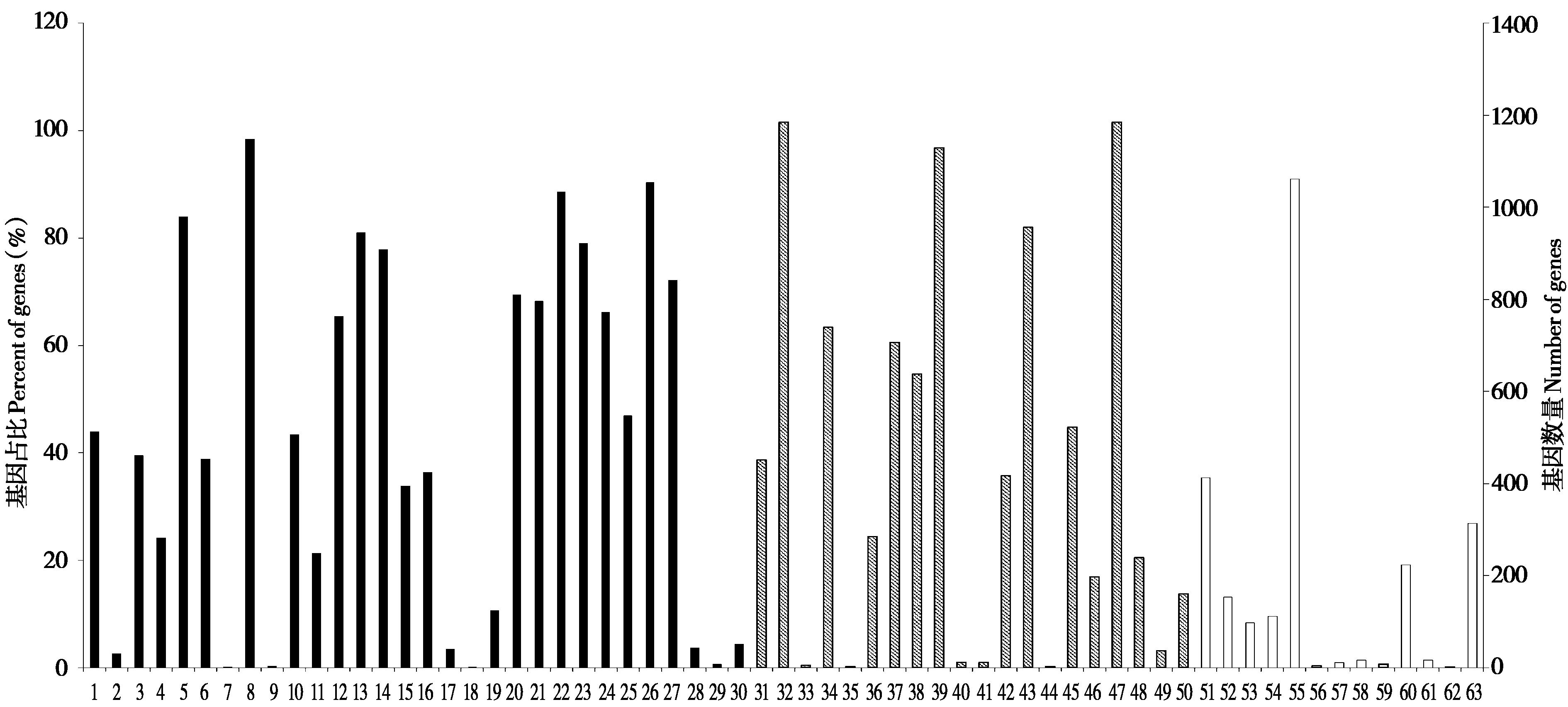
图 4 大蹄蝠微卫星分布于外显子的基因GO功能分类. 1:生殖;2:生殖细胞杀伤;3:免疫系统过程;4:行为;5:代谢;6:细胞增殖;7:碳水化合物利用;8:细胞过程;9:氮的利用;10:生殖过程;11:生物粘附;12:信号;13:多细胞生物突;14:发育过程;15:生长;16:运动;17:色素沉着;18:生物相;19:节律过程;20:生物过程的正向调节;21:生物过程负调节;22:生物过程调节;23:对刺激的反应;24:定位;25:多生物过程;26:生物调节;27:细胞组成或生物发生;28:细胞聚集;29:解毒;30:与化学突触传递有关的突触前过程;31:细胞外区;32:细胞;33:类核;34:膜;35:病毒粒子;36:细胞连接;37:膜封闭腔;38:含蛋白复合物;39:细胞器;40:其他生物;41:其他有机体部分;42:胞外区部分;43:细胞器部分;44:病毒粒子部分;45:薄膜部分;46:突触部分;47:细胞部分;48:突触;49:共质体;50:超分子络合物;51:催化活性;52:信号传感器活动;53:结构分子活性;54:转运活动;55:连接;56:抗氧化活性;57:货物受体活性;58:转换调节器活性;59:毒素活性;60:分子功能调节剂;61:被劫持的分子功能;62:分子载体活性;63:转录调节因子活性.黑色、条纹和白色区域分别代表生物过程、细胞组分和分子功能
Fig. 4 GO classifications of coding sequencings with microsatellites in the genomes of Hipposideros armiger. 1: Reproduction; 2: Cell killing; 3: Immune system process; 4: Behavior; 5: Metabolic process; 6: Cell proliferation; 7: Carbohydrate utilization; 8: Cellular process; 9: Nitrogen utilization; 10: Reproductive process; 11: Biological adhesion; 12: Signaling; 13: Multicellular organismal process; 14: Developmental process; 15: Growth; 16: Locomotion; 17: Pigmentation; 18: Biological phase; 19: Rhythmic process; 20: Positive regulation of biological process; 21: Negative regulation of biological process; 22: Regulation of biological process; 23: Response to stimulus; 24: Localization; 25: Multi-organism process; 26: Biological regulation; 27: Cellular component organization or biogenesis; 28: Cell aggregation; 29: Detoxification; 30: Presynaptic process involved in chemical synaptic transmission; 31: Extracellular region; 32: Cell; 33: Nucleoid; 34: Membrane; 35: Virion; 36: Cell junction; 37: Membrane-enclosed lumen; 38: Protein-containing complex; 39: Organelle; 40: Other organism; 41: Other organism part; 42: Extracellular region part; 43: Organelle part; 44: Virion part; 45: Membrane part; 46: Synapse part; 47: Cell part; 48: Synapse; 49: Symplast; 50: Supramolecular complex; 51: Catalytic activity; 52: Signal transducer activity; 53: Structural molecule activity; 54: Transporter activity; 55: Binding; 56: Antioxidant activity; 57: Cargo receptor activity; 58: Translation regulator activity; 59: Toxin activity; 60: Molecular function regulator; 61: Hijacked molecular function; 62: Molecular carrier activity; 63: Transcription regulator activity. The black, stripe and white area indicated biological process, cellular component and molecular function, respectively
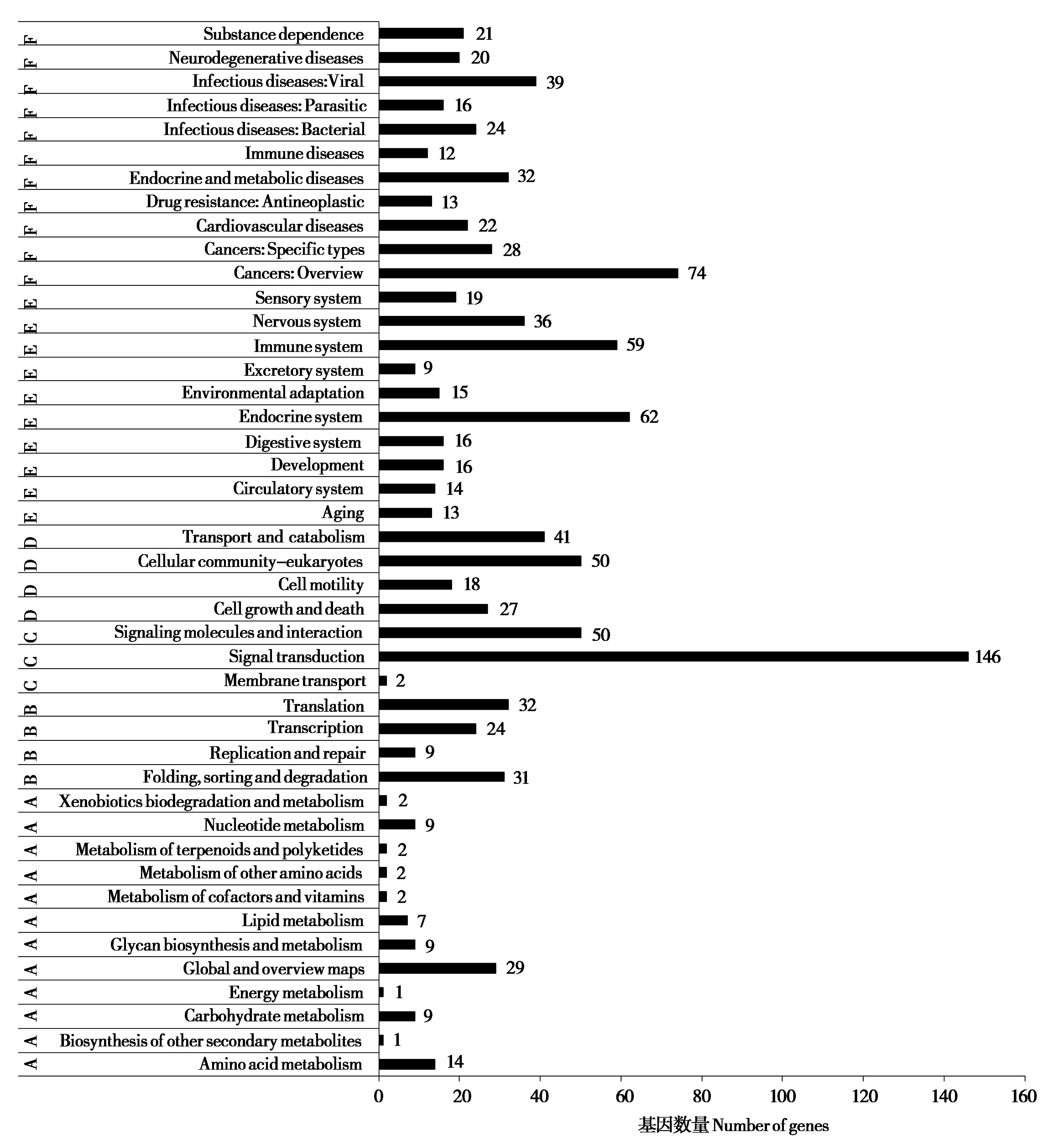
图5 大蹄蝠微卫星分布于外显子的基因KEGG富集. A:代谢;B:环境信息处理;C:遗传信息处理;D:细胞过程;E:生物体系统;F:人类疾病与药物开发
Fig. 5 The KEGG enrichment of exon microsatellites in Hipposideros armiger. A: Metabolism; B: Environmental information processing; C: Genetic information processing; D: Cell process; E: Organismal systems; F: Human diseases and drug development
| Castillo‑Lizardo M, Henneke G, Viguera E. 2014. Replication slippage of the thermophilic DNA polymerases B and D from the Euryarchaeota Pyrococcus abyssi . Frontiers in Microbiology, 5 (403): 403-403. | |
| Collaborative R. 2022. Impact of microsatellite status in early-onset colonic cancer. British Journal of Surgery, 109 (7): 632-636. | |
| Cui K, Yue B S. 2018. Distribution patterns of microsatellites in the genome of Lophophorus lhuysii . Sichuan Journal of Zoology, 37 (5): 533-540. (in Chinese) | |
| Deback C, Boutolleau D, Depienne C, Luyt C E, Bonnafous P, Gautheret‑Dejean A, Garrigue I, Agut H. 2009. Utilization of microsatellite polymorphism for differentiating herpes simplex virus type 1 strains. Journal of Clinical Microbiology, 47: 533-540. | |
| Dhyani P, Sharma B, Singh P, Masand M, Sharma R. 2020. Genome-wide discovery of microsatellite markers and, population genetic diversity inferences revealed high anthropogenic pressure on endemic populations of Trillium govanianum . Industrial Crops and Products, 154: 112698. | |
| Ding S M, Wang S P, He K, Jiang M X, Li F. 2017. Large scale analysis reveals that the genome features of simple sequence repeats are generally conserved at the family level in insects. BMC Genomics, 18: 848. | |
| Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nature Reviews Genetics, 5 (6): 435-445. | |
| Fan S G, Huang H, Liu Y, Wang P F, Zhao C, Yan L L, Qiao X T, Qiu L H. 2021. Genome‑wide identification of microsatellite and development of polymorphic SSR markers for spotted sea bass (Lateolabrax maculatus). Aquaculture Reports, 20: 100677. | |
| Fujimori S, Washio T, Higo K. 2003. A novel feature of microsatellites in plants: a distribution gradient among the direction of transcription. FEBS Letters, 554 (1): 17-22. | |
| Gao H, Liu P, Meng X H, Wang W J, Kong J. 2004. Analysis of microsatellite sequences in Chinese shrimp Fenneropenaeus chinensis genome. Oceanologia et Limnologia Sinica, 35 (5): 424-431. (in Chinese) | |
| George B, Gnanasekaran P, Jain S K, Chakraborty S. 2014. Genome wide survey and analysis of small repetitive sequences in caulimoviruses. Infection Genetics & Evolution, 27: 15-24. | |
| Hancock J M. 1996. Simple sequences in a ‘minimal’ genome. Nature Genetics, 14: 14-15. | |
| Huang J, Du L M, Li Y Z, Li W J, Zhang X Y, Yue B S. 2012. Distribution regularities of microsatellites in the Gallus gallus genome. Sichuan Journal of Zoology, 31 (3): 358-363. (in Chinese) | |
| Huntley M A, Golding G B. 2006. Selection and slippage creating serine homopolymers. Molecular Biology and Evolution, 23: 2017-2025. | |
| Kashi Y, King D G. 2006. Simple sequence repeats as advantageous mutators in evolution. Trends in Genetics, 22: 253-259. | |
| Li W J, Li Y Z, Du L M, Huang J, Shen Y M, Zhang X Y, Yue B S. 2014. Comparative analysis of microsatellite sequences distribution in the genome of giant panda and polar bear. Sichuan Journal of Zoology, 33 (6): 874-878. (in Chinese) | |
| Lin W H, Kussel E. 2012. Evolutionary pressures on simple sequence repeats in prokaryotic coding regions. Nucleic Acids Research, 40: 2399-2413. | |
| Loire E, Higuet D, Netter P, Achaz G. 2013. Evolution of coding microsatellites in primate genomes. Genome Biology and Evolution, 5: 283-295. | |
| Lu K M, Wang T J, Dong S W, Xing X M, Su W L. 2022. Research on distribution characteristics of microsatellites in Sika deer genome. Special Wild Economic Animal and Plant Research, 44 (6): 8-15, 23.(in Chinese) | |
| Lu T, Wang C, Du C, Liu S, Shen Y M, Zhang X Y, Yue B S. 2017. Distribution regularity of microsatellites in Moschus berezovskii genome. Sichuan Journal of Zoology, 36 (4): 420-424. (in Chinese) | |
| Mayer C, Leese F, Tollrian R. 2010. Genome‑wide analysis of tandem repeats in Daphnia pulex–a comparative approach. BMC Genomics, 11: 277. | |
| Miah G, Rafii M Y, Ismail M R, Puteh A B, Rahim H A, Islam K N, Latif M A. 2013. A Review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. International Journal of Molecular Science, 14: 22499-22528. | |
| Mirkin S M. 2007. Expandable DNA repeats and human disease. Nature, 447: 932-940. | |
| Nie H, Cao S S, Zhao M L, Du L F. 2017. Comparative analysis of microsatellite distributions in genomes of Boa constrictor and Protobothrops mucrosquamatus . Sichuan Journal of Zoology, 36 (6): 639-648.(in Chinese) | |
| Raghunath S. 2022. Application of Bioinformatics resources for mining of simple sequence repeats (SSRs) marker in plant genomes: an overview. Research Journal of Biotechnology, 17 (8): 136-143. | |
| Rogers J, Gibbs R A. 2014. Comparative primate genomics: emerging patterns of genome content and dynamics. Nature Reviews Genetics, 15: 347-359. | |
| Smith T H, Xie Y. 2008. A Guide to the Mammals of China. Princeton, NJ: Princeton University Press. | |
| Subramanian S, Mishra R K, Singh L. 2003. Genome‑wide analysis of microsatellite repeats in humans: their abundance and density in specific genomic regions. Genome Biology, 4 (2): 1-10. | |
| Tay W T, Behere G T, Batterham P, Heckel D G. 2010. Generation of microsatellite repeat families by RTE retrotransposons in lepidopteran genomes. BMC Evolutionary Biology, 10: 144. | |
| Tóth G, Góspóri Z, Jurka J. 2000. Microsatellites in different eukaryotic genomes: survey and analysis. Genome Research, 10: 967-981. | |
| Tu F Y, Liu J, Han W J, Huang T, Huang X F. 2018. Analysis of microsatellite distribution characteristics in the entire genome of Macaca fascicularis . Chinese Journal of Wildlife, 39 (2): 400-404. (in Chinese) | |
| Walter R, Epperson B K. 2001. Geographic pattern of genetic variation in Pinus resinosa: area of greatest diversity is not the origin of postglacial populations. Molecular Ecology, 10: 103-111. | |
| Wang J, Luo Q, Duan Q, Zhong K L, Huang X T, Zhang R Y. 2019. Screening and analysis of microsatellites in the genomes of three Sinocyclocheilus fishes. Journal of Guizhou Normal University (Natural Sciences), 37 (4): 19-24. (in Chinese) | |
| Wang Y Y, Liu X X, Dong K Z, Chen X F, Ye S H, Ma Y H. 2015. Distribution difference of microsatellite in 7 domestic animals genome. China Animal Husbandry & Veterinary Medicine, 42 (9): 2418-2426. (in Chinese) | |
| Wei L, Shao W W, Ma L, Lin Z H. 2020.Genome wide analysis of microsatellite markers based on sequenced database in two anuran species. Journal of Genetics, 99: 58. | |
| Yin W, Wang Z J, Li Q Y, Lian J M, Zhou Y, Lu B Z, Jin L J, Qiu P X, Zhang P, Zhu W B, Wen B, Huang Y J, Lin Z L, Qiu B T, Su X W, Yang H M, Zhang G J, Yan G M, Zhou Q.2016. Evolutionary trajectories of snake genes and genomes revealed by comparative analysis of five‑pacer viper. Nature Communications, 13107 (1):1-11. | |
| Zane L, Bargelloni L, Patarnello T. 2002. Strategies for microsatellite isolation: a review. Molecular Ecology, 11: 1-16. | |
| 王月月, 刘雪雪, 董坤哲, 陈潇飞, 叶绍辉, 马月辉. 2015. 7种家养动物全基因组微卫星分布的差异研究. 中国畜牧兽医, 42 (9): 2418-2426. | |
| 王军, 罗琦, 段茜, 钟凯丽, 黄晓彤, 张仁意. 2019. 3种金线鲃属鱼类基因组微卫星的筛选与分析. 贵州师范大学学报 (自然科学版), 37 (4): 19-24. | |
| 卢凯妹, 王天骄, 董世武, 邢秀梅, 苏伟林. 2022. 梅花鹿基因组微卫星分布特征研究. 特产研究, 44 (6): 8-15, 23. | |
| 卢婷, 王晨, 杜超, 刘姝, 沈咏梅, 张修月, 岳碧松. 2017. 林麝全基因组微卫星分布规律研究.四川动物, 36 (4): 420-424. | |
| 李午佼, 李玉芝, 杜联明, 黄杰, 沈咏梅, 张修月, 岳碧松. 2014. 大熊猫和北极熊基因组微卫星分布特征比较分析. 四川动物, 33 (6): 874-878. | |
| 聂虎, 曹莎莎, 赵明朗, 杜林方. 2017.红尾蚺和原矛头蝮基因组微卫星分布特征比较分析.四川动物, 36 (6): 639-648. | |
| 高焕, 刘萍, 孟宪红, 王伟继, 孔杰. 2004. 中国对虾 (Fenneropenaeus chinensis) 基因组微卫星特征分析. 海洋与湖沼, 35 (5): 424-431. | |
| 涂飞云, 刘俊, 韩卫杰, 黄挺, 黄晓凤. 2018. 食蟹猴全基因组微卫星分布特征分析. 野生动物学报, 39 (2): 400-404. | |
| 黄杰, 杜联明, 李玉芝, 李午佼, 张修月, 岳碧松. 2012. 红原鸡全基因组中微卫星分布规律研究. 四川动物, 31 (3): 358-363. | |
| 崔凯, 岳碧松. 2018. 绿尾虹雉全基因组微卫星分布规律研究. 四川动物, 37 (5): 533-540. |
| [1] | 郑凯丹, 汪巧云, 范朋飞, 韩雪松, 肖梅, 谌利民, 董正一, 张璐. 基于微卫星的欧亚水獭个体识别和遗传多样性[J]. 兽类学报, 2024, 44(2): 146-158. |
| [2] | 黄泽锋, 廖雅晴, 王晓云, 张惠光, 蔡斌, 雍凡, 崔鹏, 余文华, 吴毅. 福建省两种管鼻蝠新纪录[J]. 兽类学报, 2023, 43(4): 472-478. |
| [3] | 田新民, 张明海. 基于粪便DNA的东北马鹿遗传多样性[J]. 兽类学报, 2023, 43(1): 41-49. |
| [4] | 李彦男, 梁晓玲, 谢慧娴, 邓汶圃, 何敏怡, 余文华, 吴毅. 大墓蝠在广东省分布的再发现[J]. 兽类学报, 2023, 43(1): 122-128. |
| [5] | 赵琪, 张琪, 李浩玲, 兰月, 鄢行安, 赵贵军, 戚文华. 林麝及其近缘物种编码区微卫星分布规律及功能分析[J]. 兽类学报, 2022, 42(6): 705-715. |
| [6] | 田新民, 廉明栋, 宋雅祺, 刘小慧, 杨孟平, 陈红. 黑龙江省张广才岭北部小飞鼠的遗传多样性与种群历史动态[J]. 兽类学报, 2022, 42(4): 398-409. |
| [7] | 谢慧娴, 李彦男, 梁晓玲, 张惠光, 詹丽英, 吴毅, 余文华. 环颈蝠(Thainycteris aureocollaris)在中国分布的再发现[J]. 兽类学报, 2021, 41(4): 476-482. |
| [8] | 张沼, 张蕊, 李晓宇, 赛罕, 杨振东, 韩志庆, 鲍伟东. 内蒙古赛罕乌拉自然保护区东北马鹿种群遗传多样性与性别结构分析[J]. 兽类学报, 2021, 41(1): 42-50. |
| [9] | 王豆 许冠 王洪永 何森 卜书海 郑雪莉. 中国圈养林麝微卫星DNA多样性研究[J]. 兽类学报, 2019, 39(6): 599-607. |
| [10] | 张于光 Charlotte Hacker 张宇 薛亚东 乌力吉 代云川 罗平 谢然尼玛 Jan E Janecka 李迪强. 三江源和祁连山国家公园雪豹种群的遗传结构分析[J]. 兽类学报, 2019, 39(4): 442-449. |
| [11] | 彭乐 叶建平 朱光剑 刘志霄 张礼标. 两种同域分布蹄蝠在开阔度不同的环境中回声定位声波的可塑性[J]. 兽类学报, 2019, 39(3): 252-257. |
| [12] | 庞育兰 罗波 王漫 吴秀 冯江. 菲菊头蝠回声定位声波频率的性二态有利于性别识别[J]. 兽类学报, 2019, 39(2): 155-161. |
| [13] | 乔麦菊 冉江洪 张和民. 微卫星标记在大熊猫研究中的应用进展[J]. 兽类学报, 2019, 39(1): 103-110. |
| [14] | 闫京艳 林恭华 陈洪舰 李千 覃雯 苏建平 张同作. 青海省东部地区喜马拉雅旱獭种群遗传结构[J]. 兽类学报, 2018, 38(5): 458-466. |
| [15] | 修云芳 刘国伟 郑舒桓. 圈养小熊猫遗传多样性与种群遗传结构分析[J]. 兽类学报, 2018, 38(4): 393-401. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2