

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2022, Vol. 42 ›› Issue (4): 398-409.DOI: 10.16829/j.slxb.150629
收稿日期:2021-10-30
接受日期:2022-02-23
出版日期:2022-07-30
发布日期:2022-07-22
通讯作者:
田新民
作者简介:田新民 (1982- ),男,博士,副教授,主要从事动物分子生态学研究.
基金资助:
Xinmin TIAN( ), Mingdong LIAN, Yaqi SONG, Xiaohui LIU, Mengping YANG, Hong CHEN
), Mingdong LIAN, Yaqi SONG, Xiaohui LIU, Mengping YANG, Hong CHEN
Received:2021-10-30
Accepted:2022-02-23
Online:2022-07-30
Published:2022-07-22
Contact:
Xinmin TIAN
摘要:
小飞鼠 (Pteromys volans) 为树栖夜行滑行类啮齿动物,在森林种子传播和维持生态系统平衡等方面发挥着重要的生态学作用。本研究利用mtDNA Cytb、控制区和nDNA微卫星3种分子标记,对黑龙江省张广才岭北部的小飞鼠种群进行遗传多样性与历史动态分析。检测出Cytb全序列 (1 140 bp) 的平均单倍型多样性为0.909,平均核苷酸多样性为0.616%;控制区全序列 (1 066 bp) 的平均单倍型多样性为0.945,平均核苷酸多样性为1.698%;微卫星检测出种群平均等位基因数13.167个,观测杂合度0.727,期望杂合度0.864,近交系数0.159。结果表明,小飞鼠种群遗传多样性丰富,但存在一定程度的杂合度不足和近亲繁殖;未检测到种群近期遗传瓶颈效应,种群内无遗传分化。高比例的稀有单倍型 (≥ 60%) 、低频率等位基因与近亲繁殖,提示未来种群面临遗传多样性下降的风险,建议加大对该物种的关注和保护力度。基于Cyt b基因的系统进化关系结果表明,小飞鼠存在3个明显的遗传谱系:远东、欧亚大陆北部和日本北海道,本研究中张广才岭和大兴安岭的样本单倍型归属为远东谱系。
中图分类号:
田新民, 廉明栋, 宋雅祺, 刘小慧, 杨孟平, 陈红. 黑龙江省张广才岭北部小飞鼠的遗传多样性与种群历史动态[J]. 兽类学报, 2022, 42(4): 398-409.
Xinmin TIAN, Mingdong LIAN, Yaqi SONG, Xiaohui LIU, Mengping YANG, Hong CHEN. Genetic diversity and demographic history of Siberian flying squirrel (Pteromys volans) population in northern Zhangguangcai Mountains, Heilongjiang, China[J]. ACTA THERIOLOGICA SINICA, 2022, 42(4): 398-409.
位点 Locus | 引物序列 Primer sequence (5′ ~ 3′) | 等位基因 Allele (bp) | 退火温度 Annealing temperature (℃) |
|---|---|---|---|
| Pvol101 | F: gtcataacatcagtctttgg R: atcacaaaaaaataaataaaagtc | 100 ~ 126 | 50 |
| Pvol414 | F: aggaaataggtctagtatatgg R: tggagtatataatttttcctg | 110 ~ 142 | 50 |
| PvolE11 | F: caggactcaagggggaaaa R: gcagaagccattctactgga | 259 ~ 315 | 55 |
| PvolE52 | F: gcacaatttcagctgcttacc R: tgagctagggactacatgatatgg | 139 ~ 169 | 50 |
| PvolE63 | F: tccttactaatgtgaaccctgaca R: cagtcttcaagcacacttcct | 169 ~ 205 | 50 |
| PvolE101 | F: gggtctataatttgaaaagagaaaga R: tgttctggtggtgagaagca | 115 ~ 141 | 50 |
| Scv33 | F: ttggctcatggtttcagaga R: cccctcacttcctccatttc | 208 ~ 260 | 52 |
| Hlep591 | F: aataaatgctgctgaaacaaactc R: gctgtgcattagcctcaaag | 304 ~ 358 | 59 |
| Hlep724 | F: gccaaaccactgctatcc R: gkgrtaatcctagccacttg | 206 ~ 236 | 55 |
| Hlep801 | F: aatactkaatgsaatgtgtgcaa R: cttccatcagctcggtca | 269 ~ 301 | 55 |
| Hph172 | F: gagtccakkgccaaakgaga R: agcctggaaactaggacagtg | 158 ~ 182 | 59 |
| ScnFO354 | F: gatggacatctgaaatagtgaga R: acactgggctaaacaacaaa | 154 ~ 172 | 55 |
表1 本研究选取的12对微卫星引物信息
Table 1 Details of 12 microsatellite loci used for the study
位点 Locus | 引物序列 Primer sequence (5′ ~ 3′) | 等位基因 Allele (bp) | 退火温度 Annealing temperature (℃) |
|---|---|---|---|
| Pvol101 | F: gtcataacatcagtctttgg R: atcacaaaaaaataaataaaagtc | 100 ~ 126 | 50 |
| Pvol414 | F: aggaaataggtctagtatatgg R: tggagtatataatttttcctg | 110 ~ 142 | 50 |
| PvolE11 | F: caggactcaagggggaaaa R: gcagaagccattctactgga | 259 ~ 315 | 55 |
| PvolE52 | F: gcacaatttcagctgcttacc R: tgagctagggactacatgatatgg | 139 ~ 169 | 50 |
| PvolE63 | F: tccttactaatgtgaaccctgaca R: cagtcttcaagcacacttcct | 169 ~ 205 | 50 |
| PvolE101 | F: gggtctataatttgaaaagagaaaga R: tgttctggtggtgagaagca | 115 ~ 141 | 50 |
| Scv33 | F: ttggctcatggtttcagaga R: cccctcacttcctccatttc | 208 ~ 260 | 52 |
| Hlep591 | F: aataaatgctgctgaaacaaactc R: gctgtgcattagcctcaaag | 304 ~ 358 | 59 |
| Hlep724 | F: gccaaaccactgctatcc R: gkgrtaatcctagccacttg | 206 ~ 236 | 55 |
| Hlep801 | F: aatactkaatgsaatgtgtgcaa R: cttccatcagctcggtca | 269 ~ 301 | 55 |
| Hph172 | F: gagtccakkgccaaakgaga R: agcctggaaactaggacagtg | 158 ~ 182 | 59 |
| ScnFO354 | F: gatggacatctgaaatagtgaga R: acactgggctaaacaacaaa | 154 ~ 172 | 55 |
研究地区 Region | 地点 Location | 代码 Code | 单倍型 Haplotype | GenBank登录号 GenBank accession No. |
|---|---|---|---|---|
| 韩国Korea | 京畿道Gyeonggi-do | KGD | PV30, PV31 | EU919147, EU919146 |
| 全罗南道Jeollanam-do | KJD | PV32 ~ PV36 | EU919142 ~ EU919145, EU919148 ~ EU919155 | |
| 中国China | 黑龙江省Heilongjiang Province | CHJ | PV41 ~ PV45 | FJ360736 ~ FJ360740 |
俄罗斯东部 Eastern Russia | 兴凯湖Khanka Lake (滨海边疆区Primorsky Krai) | RKL | PV11, PV17 | AB164656, AB164662 |
| 乌苏里江Ussuri River (滨海边疆区Primorsky Krai) | RUS | PV13 | AB164658 | |
| 松花江Sungari River (滨海边疆区Primorsky Krai) | RSU | PV08 | AB164653 | |
| 苏昌河Suchan River (滨海边疆区Primorsky Krai) | RSH | PV22 | AB164667 | |
俄罗斯西部 Western Russia | 列宁格勒州Leningrad | RLE | PV14, PV03 | AB164659, AB164648 |
| 卡累利阿共和国Karlia | RKA | PV10 | AB164655 | |
| 诺夫哥罗德州Novgorod | RNO | PV09 | AB164654 | |
| 莫斯科州Moskovskaya | RMO | ZMMU: S195971 | KT962998 | |
| 基洛夫州Kirov | RKI | PV25, PV40 | AB164670, EU919156 | |
| 彼尔姆边疆区Permskiy | RPE | PV03 | AB164648 | |
| 乌拉尔山脉Ural Mountains | RUM | Pt1, Pt6, Pt8 | KR063240 ~ KR063244 | |
| 阿尔泰共和国Altai | RAL | PV24 | AB164669 | |
| 克拉斯诺亚尔斯克边疆区Kansnoyarsk | RKY | PV23, PV02 | AB164668, AB164479 | |
| 萨彦岭山脉Sayan Mountains | RSM | PV04 | AB164649 | |
俄罗斯中西伯利亚 Central Siberian of Russia | 伊尔库茨克州Irkutsk | RIR | PV21 | AB164666 |
| 雅库特共和国Yakutia | RYA | PV12, PV15 ~ PV19 | AB164657, AB164660 ~ AB164664 | |
| 布里亚特共和国Buryatia | RBU | PV07 | AB164652 | |
| 赤塔州Chitinskajaoblast | RCH | PV37 ~ PV39 | EU919157 ~ EU919160 | |
| 阿穆尔州Amur | RAM | PV01 | AB164478 | |
| 阿纳德尔Anadyr (楚科奇自治区Chukotskiy) | RAN | PV20 | AB164665 | |
| 蒙古Mongolia | 蒙古北部North Mongolia | MON | PV05, PV06 | AB164650, AB164651 |
| 日本Japan | 北海道Hokkaido | JHO | PV26 ~ PV29 | AB164671 ~ AB164674, AB097683, AB023910 |
表2 GenBank下载的小飞鼠Cyt b基因信息
Table 2 Mitochondrial cytochrome b gene of Siberian flying squirrel from GenBank
研究地区 Region | 地点 Location | 代码 Code | 单倍型 Haplotype | GenBank登录号 GenBank accession No. |
|---|---|---|---|---|
| 韩国Korea | 京畿道Gyeonggi-do | KGD | PV30, PV31 | EU919147, EU919146 |
| 全罗南道Jeollanam-do | KJD | PV32 ~ PV36 | EU919142 ~ EU919145, EU919148 ~ EU919155 | |
| 中国China | 黑龙江省Heilongjiang Province | CHJ | PV41 ~ PV45 | FJ360736 ~ FJ360740 |
俄罗斯东部 Eastern Russia | 兴凯湖Khanka Lake (滨海边疆区Primorsky Krai) | RKL | PV11, PV17 | AB164656, AB164662 |
| 乌苏里江Ussuri River (滨海边疆区Primorsky Krai) | RUS | PV13 | AB164658 | |
| 松花江Sungari River (滨海边疆区Primorsky Krai) | RSU | PV08 | AB164653 | |
| 苏昌河Suchan River (滨海边疆区Primorsky Krai) | RSH | PV22 | AB164667 | |
俄罗斯西部 Western Russia | 列宁格勒州Leningrad | RLE | PV14, PV03 | AB164659, AB164648 |
| 卡累利阿共和国Karlia | RKA | PV10 | AB164655 | |
| 诺夫哥罗德州Novgorod | RNO | PV09 | AB164654 | |
| 莫斯科州Moskovskaya | RMO | ZMMU: S195971 | KT962998 | |
| 基洛夫州Kirov | RKI | PV25, PV40 | AB164670, EU919156 | |
| 彼尔姆边疆区Permskiy | RPE | PV03 | AB164648 | |
| 乌拉尔山脉Ural Mountains | RUM | Pt1, Pt6, Pt8 | KR063240 ~ KR063244 | |
| 阿尔泰共和国Altai | RAL | PV24 | AB164669 | |
| 克拉斯诺亚尔斯克边疆区Kansnoyarsk | RKY | PV23, PV02 | AB164668, AB164479 | |
| 萨彦岭山脉Sayan Mountains | RSM | PV04 | AB164649 | |
俄罗斯中西伯利亚 Central Siberian of Russia | 伊尔库茨克州Irkutsk | RIR | PV21 | AB164666 |
| 雅库特共和国Yakutia | RYA | PV12, PV15 ~ PV19 | AB164657, AB164660 ~ AB164664 | |
| 布里亚特共和国Buryatia | RBU | PV07 | AB164652 | |
| 赤塔州Chitinskajaoblast | RCH | PV37 ~ PV39 | EU919157 ~ EU919160 | |
| 阿穆尔州Amur | RAM | PV01 | AB164478 | |
| 阿纳德尔Anadyr (楚科奇自治区Chukotskiy) | RAN | PV20 | AB164665 | |
| 蒙古Mongolia | 蒙古北部North Mongolia | MON | PV05, PV06 | AB164650, AB164651 |
| 日本Japan | 北海道Hokkaido | JHO | PV26 ~ PV29 | AB164671 ~ AB164674, AB097683, AB023910 |

图2 小飞鼠种群14个Cytb单倍型变异位点.N:具有相同单倍型的个体数量
Fig. 2 The variable sites of 14 cytochrome b haplotypes identified in Siberian flying squirrel population.N: Number of individuals with each haplotype
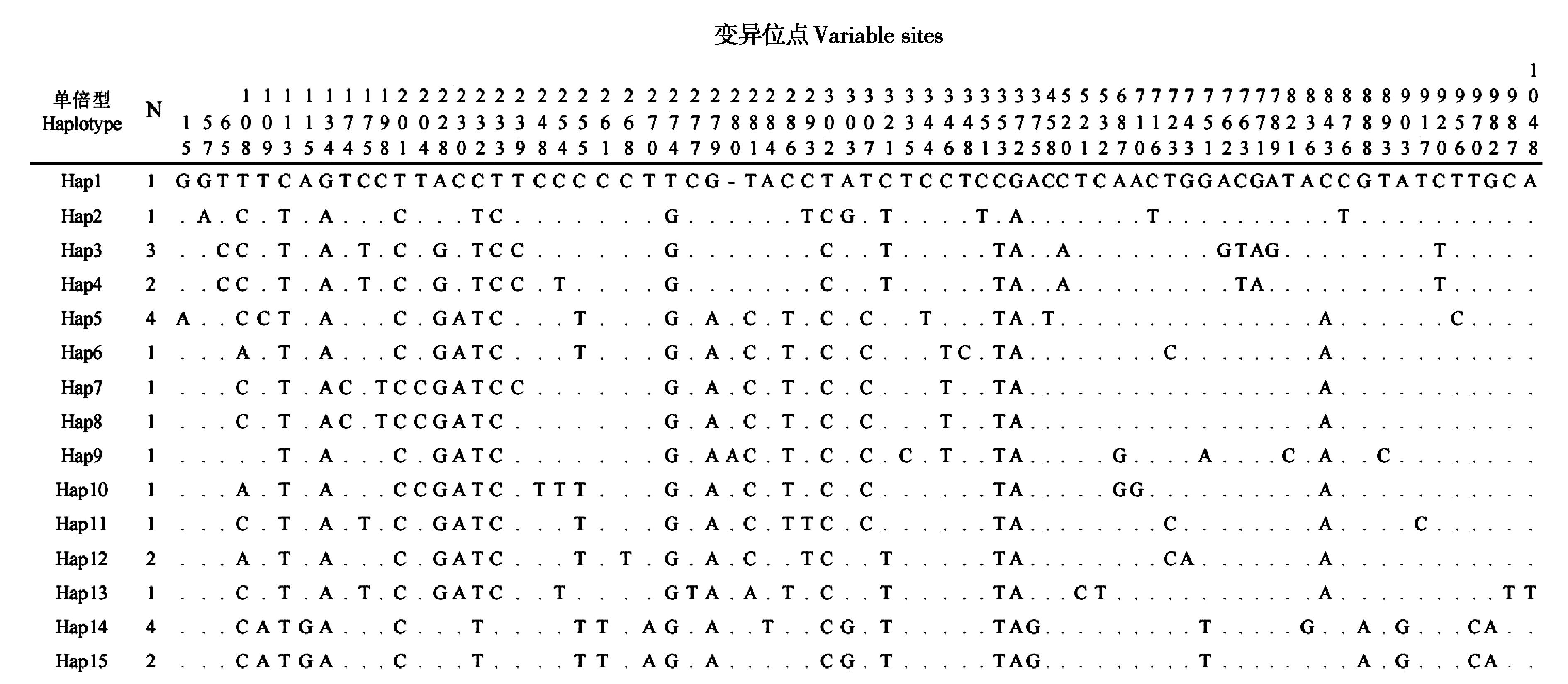
图3 小飞鼠种群15个控制区单倍型变异位点.N:具有相同单倍型的个体数量
Fig. 3 The variable sites of 15 control region haplotypes identified in Siberian flying squirrel population.N: Number of individuals with each haplotype
项目 Item | 序列数 Number of sequences | 变异位点数 Number of variable sites | 单倍型个数 Number of haplotypes | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity (%) | Tajima’s D | Fu’s Fs |
|---|---|---|---|---|---|---|---|
| Cyt b | 29 | 34 | 14 | 0.909 ± 0.031 | 0.616 ± 0.065 | -0.694 | -1.108 |
| CR | 26 | 72 | 15 | 0.945 ± 0.024 | 1.698 ± 0.068 | -0.307 | 1.376 |
表3 基于Cytb与控制区序列的种群遗传多样性分析
Table 3 Analysis of population genetic diversity based on cytochrome b (Cytb) and control region (CR)
项目 Item | 序列数 Number of sequences | 变异位点数 Number of variable sites | 单倍型个数 Number of haplotypes | 单倍型多样性 Haplotype diversity | 核苷酸多样性 Nucleotide diversity (%) | Tajima’s D | Fu’s Fs |
|---|---|---|---|---|---|---|---|
| Cyt b | 29 | 34 | 14 | 0.909 ± 0.031 | 0.616 ± 0.065 | -0.694 | -1.108 |
| CR | 26 | 72 | 15 | 0.945 ± 0.024 | 1.698 ± 0.068 | -0.307 | 1.376 |
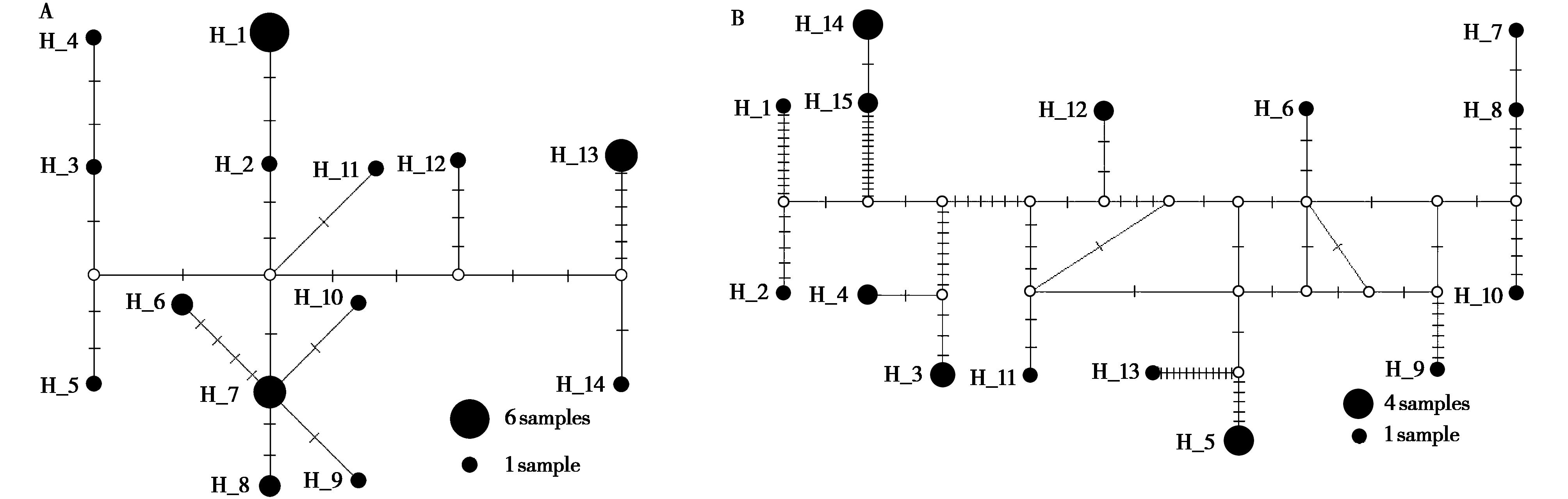
图4 基于Cytb (A) 与控制区 (B) 序列构建的单倍型网络图. 圆形面积代表单倍型频次,空心点代表缺失的单倍型
Fig. 4 Median-joining network of haplotypes based on Cytb (A) and control region (B). The size of each circle represents the frequency of haplotypes, and small hollow dot represents missing haplotype
位点 Locus | Na | Ne | PIC | Ho | He | Fis | PHW |
|---|---|---|---|---|---|---|---|
| Pvol10 | 13 | 8.899 | 0.877 | 0.759 | 0.888 | 0.145 | 0.045* |
| Pvol41 | 13 | 8.245 | 0.867 | 0.897 | 0.879 | -0.020 | 0.948 |
| PvolE1 | 18 | 10.513 | 0.908 | 0.414 | 0.905 | 0.543 | 0.000** |
| PvolE5 | 14 | 9.043 | 0.880 | 0.724 | 0.889 | 0.186 | 0.008** |
| PvolE6 | 14 | 5.644 | 0.806 | 0.828 | 0.823 | -0.006 | 0.158 |
| PvolE10 | 11 | 5.986 | 0.806 | 0.552 | 0.833 | 0.338 | 0.000** |
| Scv3 | 15 | 6.050 | 0.868 | 0.586 | 0.835 | 0.298 | 0.000** |
| Hlep59 | 17 | 10.922 | 0.901 | 0.966 | 0.908 | -0.063 | 0.669 |
| Hlep72 | 12 | 7.897 | 0.863 | 0.931 | 0.873 | -0.066 | 0.814 |
| Hlep80 | 12 | 5.207 | 0.819 | 0.517 | 0.808 | 0.360 | 0.000** |
| Hph17 | 11 | 7.543 | 0.851 | 0.793 | 0.867 | 0.086 | 0.260 |
| ScnFO-35 | 8 | 6.622 | 0.829 | 0.759 | 0.849 | 0.106 | 0.112 |
| 平均值Mean | 13.167 | 7.714 | 0.856 | 0.727 | 0.864 | 0.159 | 0.000** |
表4 基于12个微卫星位点的种群遗传多样性分析
Table 4 Analysis of population genetic diversity based on 12 microsatellite loci
位点 Locus | Na | Ne | PIC | Ho | He | Fis | PHW |
|---|---|---|---|---|---|---|---|
| Pvol10 | 13 | 8.899 | 0.877 | 0.759 | 0.888 | 0.145 | 0.045* |
| Pvol41 | 13 | 8.245 | 0.867 | 0.897 | 0.879 | -0.020 | 0.948 |
| PvolE1 | 18 | 10.513 | 0.908 | 0.414 | 0.905 | 0.543 | 0.000** |
| PvolE5 | 14 | 9.043 | 0.880 | 0.724 | 0.889 | 0.186 | 0.008** |
| PvolE6 | 14 | 5.644 | 0.806 | 0.828 | 0.823 | -0.006 | 0.158 |
| PvolE10 | 11 | 5.986 | 0.806 | 0.552 | 0.833 | 0.338 | 0.000** |
| Scv3 | 15 | 6.050 | 0.868 | 0.586 | 0.835 | 0.298 | 0.000** |
| Hlep59 | 17 | 10.922 | 0.901 | 0.966 | 0.908 | -0.063 | 0.669 |
| Hlep72 | 12 | 7.897 | 0.863 | 0.931 | 0.873 | -0.066 | 0.814 |
| Hlep80 | 12 | 5.207 | 0.819 | 0.517 | 0.808 | 0.360 | 0.000** |
| Hph17 | 11 | 7.543 | 0.851 | 0.793 | 0.867 | 0.086 | 0.260 |
| ScnFO-35 | 8 | 6.622 | 0.829 | 0.759 | 0.849 | 0.106 | 0.112 |
| 平均值Mean | 13.167 | 7.714 | 0.856 | 0.727 | 0.864 | 0.159 | 0.000** |
物种 Species | 研究地区Region | Cyt b | 控制区 Control region | 微卫星 Microsatellite | 文献Reference | |||
|---|---|---|---|---|---|---|---|---|
| Hd | Pi (%) | Hd | Pi (%) | Ho | He | |||
小飞鼠 Pteromys volans | 中国黑龙江省 Heilongjiang, China | 0.909 | 0.616 | 0.945 | 1.698 | 0.727 | 0.864 | 本研究 This study |
爱沙尼亚 Estonia | 0.000 | 0.000 | 0.305 | 0.048 | 0.608 | 0.764 | ||
芬兰 Finland | 0.125 | 0.027 | 0.642 | 0.156 | 0.410 ~ 0.772 | 0.430 ~ 0.748 | ||
俄罗斯 Russia | 0.222 ~ 1.000 | 0.097 ~ 0.950 | 0.700 | 0.127 | — | — | ||
日本北海道 Hokkaido, Japan | 1.000 | 0.219 | — | — | — | — | ||
中国黑龙江省 Heilongjiang, China | 1.000 | 0.614 | — | — | — | — | ||
| 韩国Korea | 0.846 | 0.616 | — | — | — | — | ||
北美飞鼠 Glaucomys sabrinus | 美国阿拉斯加州 Alaska, American | — | — | — | — | 0.247 ~ 0.624 | 0.128 ~ 0.644 | |
美国布拉克山 Black Hills, American | — | — | — | — | 0.36 | 0.35 | ||
南方飞鼠 Glaucomys volans | 美国南卡罗来纳州 South Carolina, American | — | — | — | — | 0.589 | 0.607 | |
表5 基于mtDNA和微卫星位点的鼯鼠科主要物种种群遗传多样性参数比较
Table 5 Comparison of population genetic diversity at mtDNA and microsatellite loci in the Pteromyidae
物种 Species | 研究地区Region | Cyt b | 控制区 Control region | 微卫星 Microsatellite | 文献Reference | |||
|---|---|---|---|---|---|---|---|---|
| Hd | Pi (%) | Hd | Pi (%) | Ho | He | |||
小飞鼠 Pteromys volans | 中国黑龙江省 Heilongjiang, China | 0.909 | 0.616 | 0.945 | 1.698 | 0.727 | 0.864 | 本研究 This study |
爱沙尼亚 Estonia | 0.000 | 0.000 | 0.305 | 0.048 | 0.608 | 0.764 | ||
芬兰 Finland | 0.125 | 0.027 | 0.642 | 0.156 | 0.410 ~ 0.772 | 0.430 ~ 0.748 | ||
俄罗斯 Russia | 0.222 ~ 1.000 | 0.097 ~ 0.950 | 0.700 | 0.127 | — | — | ||
日本北海道 Hokkaido, Japan | 1.000 | 0.219 | — | — | — | — | ||
中国黑龙江省 Heilongjiang, China | 1.000 | 0.614 | — | — | — | — | ||
| 韩国Korea | 0.846 | 0.616 | — | — | — | — | ||
北美飞鼠 Glaucomys sabrinus | 美国阿拉斯加州 Alaska, American | — | — | — | — | 0.247 ~ 0.624 | 0.128 ~ 0.644 | |
美国布拉克山 Black Hills, American | — | — | — | — | 0.36 | 0.35 | ||
南方飞鼠 Glaucomys volans | 美国南卡罗来纳州 South Carolina, American | — | — | — | — | 0.589 | 0.607 | |
| Arbogast B S, Browne R A, Weig P D, Kenagy G J. 2005. Conservation genetics of endangered flying squirrels (Glaucomys) from the Appalachian Mountains of eastern North America. Animal Conservation, 8 (2): 123-133. | |
| Balakrishnan C N, Monfort S L, Gaur A, Sing L, Sorenson M D. 2003. Phylogeography and conservation genetics of Eld’s deer (Cervus eldi). Molecular Ecology, 12 (1): 1-10. | |
| Bandelt H J, Forster P, Röhl A. 1999. Median-joining networks for inferring intraspecific phylogenics. Molecular Biology Evolution, 16 (1): 37-48. | |
| Bellemain E, Swenson J E, Tallmon D, Brunberg S, Taberlet P. 2005. Estimating population size of elusive animals with DNA from hunter-collected feces: four methods for brown bears. Conservation Biology, 19 (1): 150-161. | |
| Bidlack A L, Cook J K. 2002. A nuclear perspective on endemism in northern flying squirrels (Glaucomys sabrinus) of the Alexander Archipelago, Alaska. Conservation Genetics, 3 (3): 247-259. | |
| Botstein D, White R L, Skolnick M, Davis R W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32 (3): 314-331. | |
| Chen L. 2018. Study on the insulation mechanism of tree cavities, as exampled from wintering cavity of Pteromys volans in northern Greater Hingan Mt’s. Ph.D thesis. Harbin: Northeast Forestry University. (in Chinese) | |
| Chen S Y, Holyoak M, Liu H, Bao H, Ma Y J, Dou H J, Jiang G S. 2022. Effects of spatially heterogeneous warming on gut microbiota, nutrition and gene flow of a heat‑sensitive ungulate population. Science of the Total Environment, 806: 150537. | |
| Crochet P A, Desmarais E. 2000. Slow rate of evolution in the mitochondrial control region of gulls (Aves: Laridae). Molecular Biology and Evolution, 17 (1): 1797-1806. | |
| Darriba D, Taboada G L, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods, 9 (8): 772. | |
| Drummond A J, Suchard M A, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution, 29 (8): 1969-1973. | |
| Earl D A, vonHoldt B M. 2012. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4 (2): 359-361. | |
| Ellegren H. 2004. Microsatellites: simple sequences with complex evolution. Nature Reviews Genetics, 5 (6): 435-445. | |
| Frankham R, Ballou J D, Briscoe D A. 2010. Introduction to Conservation Genetics. New York: Cambridge University Press, 235-237. | |
| Grant W S, Bowen B W. 1998. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of Heredity, 89 (5): 415-426. | |
| Haig S M, Ballou J D, Derrickson S R. 1990. Management options for preserving genetic diversity: reintroduction of Guam rails to the wild. Conservation Biology, 4 (3): 290-300. | |
| Hale M L, Bevan R, Wolff K. 2001. New polymorphic microsatellite markers for the red squirrel (Sciurus vulgaris) and their applicability to the grey squirrel (S .carolinensis). Molecular Ecology Notes, 1 (1/2): 47-49. | |
| Hanski I K, Stevens P C, Ihalempiä P, Selonen V. 2000. Home-range size, movements, and nest-site use in the Siberian flying squirrel, Pteromys volans . Journal of Mammalogy, 81 (3): 798-809. | |
| Hurme E, Monkkonen M, Sippola A L, Ylinen H, Pentinsaari M. 2007. Role of the Siberian flying squirrel as an umbrella species for biodiversity in northern boreal forests. Ecological Indicators, 8 (3): 246-255. | |
| Jiang Z G, Jiang J P, Wang Y Z, Zhang E, Zhang Y Y, Li L L, Xie F, Cai B, Cao L, Zheng G M, Dong L, Zhang Z W, Ding P, Luo Z H, Ding C Q, Ma Z J, Tang S H, Cao W X, Li C W, Hu H J, Ma Y, Wu Y, Wang Z J, Wang Y, Wang B, Li C, Song X L, Cai L, Zang C X, Zeng Y, Meng Z B, Fang H X, Ping X G. 2016. Red list of China’s vertebrates. Biodiversity Science, 24 (5): 501-551. (in Chinese) | |
| Jiang Z G, Ma Y, Wu Y, Wang Y X, Zhou K Y, Liu S Y, Feng Z J. 2015. China’s Mammal Diversity and Geographic Distribution . Beijing: China Science Press, 276. (in Chinese) | |
| Jumpa S, Dawson D A, Horsburgh G J, Walton C. 2015. Conserved microsatellite markers of high cross-species utility for flying, ground and tree squirrels. Conservation Genetics Resources, 7 (2): 599-603. | |
| Kiesow A M, Monroe E M, Britten H B. 2012. Genetic structure of the arboreal squirrels (Glaucomys sabrinus and Tamiasciurus hudsonicus) in the North American Black Hills. Canadian Journal of Zoology, 90 (9): 1191-1200. | |
| Kiesow A M, Wallace L E, Britten H B. 2011. Characterization and isolation of five microsatellite loci in northern flying squirrels, Glaucomys sabrinus (Sciuridae, Rodentia). Western North American Naturalist, 71 (4): 553-556. | |
| Krojerová-Prokešová J, Barančeková M, Voloshina I, Myslenkov A, Lamka J, Koubek P. 2013. Dybowski’s sika deer (Cervus nippon hortulorum): genetic divergence between natural Primorian and introduced Czech populations. Journal of Heredity, 104 (3): 312-326. | |
| Lade J A, Murray N D, Marks C A, Robinson N A. 1996. Microsatellite differentiation between Phillip Island and mainland Australian populations of the red fox Vulpes vulpes . Molecular Ecology, 5 (1): 81-87. | |
| Lampila S, Kvist L, Wistbacka R, Orell M. 2009. Genetic diversity and population differentiation in the endangered Siberian flying squirrel (Pteromys volans) in a fragmented landscape. European Journal of Wildlife Research, 55 (4): 397-406. | |
| Larkin M A, Blackshields G, Brown N P, Chenna R, McGettigan P A, McWilliam H, Valentin F, Wallace I M, Wilm A, Lopez R. 2007. Clustal W and Clustal X version 2.0. Bioinformatics, 23 (11): 2947-2948. | |
| Lee M Y, Lissovsky A A, Park S K, Obolenskaya E V, Dokuchaev N E, Zhang Y P, Yu L, Kim Y J, Voloshina I, Myslenkov A, Choi T Y, Min M S, Lee H. 2008b. Mitochondrial cytochrome b sequence variations and population structure of Siberian chipmunk (Tamias sibiricus) in northeastern Asia and population substructure in South Korea. Molecules and Cells, 26 (6): 566-575. | |
| Lee M Y, Park S K, Hong Y J, Kim Y J, Voloshina I, Myslenkov A, Saveljev A P, Choi T Y, Piao R Z, An J H, Lee M H, Lee H, Min M S. 2008a. Mitochondrial genetic diversity and phylogenetic relationships of Siberian flying squirrel (Pteromys volans) populations. Animal Cells and Systems, 12 (4): 269-277. | |
| Li W P, Bao H, Zhang M H. 2017. Habitat analysis and design of potential ecological corridors for Amur tiger in Northeastern China. Acta Theriologica Sinica, 37 (4): 317-326. (in Chinese) | |
| Librado P, Rozas J. 2009. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics, 25 (11): 1451-1452. | |
| Lim S J, Kim K Y, Kim H R, Chae H M, Park Y C. 2018. Phylogenetic position of the Siberian flying squirrel Pteromys volans based on complete mitochondrial genome sequences. Mitochondrial DNA Part B-Resources, 3 (2): 601-602. | |
| Ma Y, Li H L, He J, Zhao Y M, Yang H Q, Lu L, Liu Q Y. 2019. Genetic diversity and phylogenetic relationships based on mtDNA control region sequences of Marmota himalayana . Acta Theriologica Sinica, 39 (3): 285-294. (in Chinese) | |
| Nakama S, Yanagawa H. 2009. Characteristics of tree cavities used by Pteromys volans orii in winter. Mammal Study, 34 (3): 161-164. | |
| Neigel J E, Avise J C. 1993. Application of a random walk model to geographic distributions of animal mitochondrial DNA variation. Genetics, 135 (4): 1209-1220. | |
| Nummert G, Aaspõllu A, Kuningas K, Timm U, Hanski I K, Maran T. 2020. Genetic diversity in Siberian flying squirrel (Pteromys volans) in its western frontier with a focus on the Estonian population. Mammal Research, 65 (4): 767-778. | |
| Oshida T, Abramov A, Yanagawa H, Masuda R. 2005. Phylogeography of the Russian flying squirrel (Pteromys volans): implication of refugia theory in arboreal small mammal of Eurasia. Molecular Ecology, 14 (4): 1191-1196. | |
| Painter J N, Selonen V, Hanski I K. 2004. Microsatellite loci for the Siberian flying squirrel, Pteromys volans . Molecular Ecology Notes, 4 (1): 119-121. | |
| Park S D E. 2001. Trypanotolerance in west african cattle and the population genetic effects of selection. Ph.D thesis. Dublin: University of Dublin. | |
| Peakall R, Smouse P E. 2006. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6 (1): 288-295. | |
| Pérez-Espona S, Pérez-Barbería F J, Mcleod J E, Jiggins C D, Gordon I J, Pemberton J M. 2008. Landscape features affect gene flow of Scottish Highland red deer (Cervus elaphus). Molecular Ecology, 17 (4): 981-996. | |
| Piry S, Luikart G, Cornuet J M. 1999. BOTTLENECK: a computer program for detecting recent reductions in the effective size using allele frequency data. Journal of Heredity, 90 (4): 502-503. | |
| Pritchard J K, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics, 155 (2): 945-959. | |
| Raymond M, Rousset F. 1995. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. Journal of Heredity, 86 (3): 248-249. | |
| Ren P, Gong K, Bao Y X, Huang X X, Zhou X, Han J Q. 2017. Parentage verification and mating system of Chinese muntjac (Muntiacus reevesi) based on fecal DNA. Acta Ecologica Sinica, 37 (20): 6933-6944. (in Chinese) | |
| Ryu S H, Kwak M J, Hwang U W. 2013. Complete mitochondrial genome of the Eurasian flying squirrel Pteromys volans (Sciuromorpha, Sciuridae) and revision of rodent phylogeny. Molecular Biology Reports, 40 (2): 1917-1926. | |
| Selonen V, Hanski I K. 2004. Young flying squirrels (Pteromys volans) dispersing in fragmented forests. Behavioral Ecology, 15 (4): 564-571. | |
| Selonen V, Hanski I K. 2012. Dispersing Siberian flying squirrels (Pteromys volans) locate preferred habitats in fragmented landscapes. Canadian Journal of Zoology, 90 (7): 885-892. | |
| Selonen V, Painter J N, Hanski I K. 2005. Microsatellite variation in the Siberian flying squirrel in Finland. Annales Zoologici Fennici, 42 (5): 505-511. | |
| Singh V K, Mangalam A K, Dwivedi S, Naik S. 1998. Primer premier: program for design of degenerate primers from a protein sequence. Biotechniques, 24 (2): 318-319. | |
| Tian X M. 2021. Studies on population genetics and factors driving differentiation of Cervus canadensis xanthopygus . Ph.D thesis. Harbin: Northeast Forestry University. (in Chinese) | |
| Todd R. 2000. Microsatellite loci in the Eurasian red squirrel, Sciurus vulgaris L. Molecular Ecology, 9 (12): 2165-2166. | |
| van Oosterhout C, Hutchinson W F, Wills D P M, Shipley P. 2004. MICRO - CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes, 4 (3): 535-538. | |
| Wang Z B. 2000. The state forestry administration makes No. 7 -state protection beneficial or have important economic and scientific research value of terrestrial wildlife list. Chinese Journal of Wildlife, 21 (5): 49-82. (in Chinese) | |
| Wei F W, Ma T X, Hu Y B. 2021. Research advances and perspectives of conservation genetics of threatened mammals in China. Acta Theriologica Sinica, 41 (5): 571-580. (in Chinese) | |
| Weir B S, Cockerham C C. 1984. Estimating F-statistics for the analysis of population structure. Evolution, 38 (6): 1358-1370. | |
| Yalkovskaya L E, Bol’shakov V N, Sibiryakov P A, Borodin A V. 2015. Phylogeography of the Siberian flying squirrel (Pteromys volans L., 1785) and the history of the formation of the modern species range: new data. Doklady Biochemistry and Biophysics, 462 (1): 181-184. | |
| Yuasa T, Nagata J, Hamasaki S, Tsuruga T, Furubayashi K. 2007. The impact of habitat fragmentation on genetic structure of Japanese sika deer (Cervus nippon) in southern Kantoh, revealed by mitochondrial D-loop sequences. Ecological Research, 22 (1): 97-106. | |
| Zhang Y G, Hacker C, Zhang Y, Xue Y D, Wu L J, Dai Y C, Luo P, Xierannima, Janecka J E, Li D Q. 2019. An analysis of genetic structure of snow leopard populations in Sanjiangyuan and Qilianshan National Parks. Acta Theriologica Sinica, 39 (4): 442-449. (in Chinese) | |
| Zittlau K A, Davis C S, Strobeck C. 2000. Characterization of microsatellite loci in northern flying squirrels (Glaucomys sabrinus). Molecular Ecology, 9 (6): 826-827. | |
| Zorigul I, Shamshinur M, Buweihailiqiemu A, Arzigul S, Subinur E, Mahmut H. 2019. Influence of environmental factors on genetic diversity of Gazella subgotturosa in Xinjiang, China. Acta Theriologica Sinica, 39 (3): 276-284. (in Chinese) | |
| 马英, 李海龙, 何建, 赵延梅, 杨汉青, 鲁亮, 刘起勇. 2019. 喜马拉雅旱獭线粒体DNA控制区遗传多样性及系统发育. 兽类学报, 39 (3): 285-294. | |
| 王志宝. 2000. 国家林业局令第七号:国家保护的有益的或者有重要经济、科学研究价值的陆生野生动物名录. 野生动物学报, 21 (5): 49-82. | |
| 田新民. 2021. 东北马鹿种群遗传学及遗传分化驱动因素研究. 哈尔滨: 东北林业大学博士学位论文. | |
| 任鹏, 龚堃, 鲍毅新, 黄相相, 周晓, 韩金巧. 2017. 基于粪便DNA的小麂亲权鉴定和婚配制研究. 生态学报, 37 (20): 6933-6944. | |
| 李维平, 包衡, 张明海. 2017. 中国东北虎栖息地分布与潜在生态廊道构建. 兽类学报, 37 (4): 317-326. | |
| 佐日古丽·伊斯马伊力, 先木西努·莫合德, 布威海丽且姆·阿巴拜科日, 阿尔孜古力·沙塔尔, 苏比奴尔·艾力, 马合木提·哈力克. 2019. 环境因子对新疆鹅喉羚群体遗传多样性的影响. 兽类学报, 39 (3): 276-284. | |
| 张于光, Hacker C, 张宇, 薛亚东, 乌力吉, 代云川, 罗平, 谢然尼玛, Janecka J E, 李迪强. 2019. 三江源和祁连山国家公园雪豹种群的遗传结构分析. 兽类学报, 39 (4): 442-449. | |
| 陈亮. 2018. 树洞保温机制研究:以大兴安岭北部小飞鼠 (Pteromys volans) 越冬树洞为例. 哈尔滨: 东北林业大学博士学位论文. | |
| 蒋志刚, 马勇, 吴毅, 王应祥, 周开亚, 刘少英, 冯祚建. 2015. 中国哺乳动物多样性及地理分布. 北京: 科学出版社, 276. | |
| 蒋志刚, 江建平, 王跃招, 张鹗, 张雁云, 李立立, 谢锋, 蔡波, 曹亮, 郑光美, 董路, 张正旺, 丁平, 罗振华,丁长青, 马志军, 汤宋华, 曹文宣, 李春旺, 胡慧建, 马勇, 吴毅, 王应祥, 周开亚, 刘少英, 陈跃英, 李家堂, 冯祚建, 王燕, 王斌, 李成, 宋雪琳, 蔡蕾, 臧春鑫, 曾岩, 孟智斌, 方红霞, 平晓鸽. 2016. 中国脊椎动物红色名录. 生物多样性, 24 (5): 501-551. | |
| 魏辅文, 马天笑, 胡义波. 2021. 中国濒危兽类保护遗传学研究进展与展望. 兽类学报, 41 (5): 571-580. |
| [1] | 郑凯丹, 汪巧云, 范朋飞, 韩雪松, 肖梅, 谌利民, 董正一, 张璐. 基于微卫星的欧亚水獭个体识别和遗传多样性[J]. 兽类学报, 2024, 44(2): 146-158. |
| [2] | 赛青高娃, 王子涵, 李全邦, 王东, 连新明. 藏羚交配群成员间亲缘关系及遗传多样性分析[J]. 兽类学报, 2024, 44(2): 159-170. |
| [3] | 邵伟伟, 乔芬, 蔡玮, 林植华, 韦力. 大蹄蝠全基因组微卫星分布特征分析[J]. 兽类学报, 2023, 43(2): 182-192. |
| [4] | 田新民, 张明海. 基于粪便DNA的东北马鹿遗传多样性[J]. 兽类学报, 2023, 43(1): 41-49. |
| [5] | 赵琪, 张琪, 李浩玲, 兰月, 鄢行安, 赵贵军, 戚文华. 林麝及其近缘物种编码区微卫星分布规律及功能分析[J]. 兽类学报, 2022, 42(6): 705-715. |
| [6] | 成若通, 陈一博, 孟祥琼, 陈家瑞, 魏青. 藏羚Y染色体雄性特异区遗传多样性[J]. 兽类学报, 2022, 42(5): 609-614. |
| [7] | 王东辉, 刘玉良, 沈富军, 蔡志刚, 安俊辉, 侯蓉. 圈养大熊猫冷冻精液对其种群遗传多样性的作用[J]. 兽类学报, 2022, 42(3): 261-269. |
| [8] | 魏辅文, 马天笑, 胡义波. 中国濒危兽类保护遗传学研究进展与展望[J]. 兽类学报, 2021, 41(5): 571-580. |
| [9] | 张沼, 张蕊, 李晓宇, 赛罕, 杨振东, 韩志庆, 鲍伟东. 内蒙古赛罕乌拉自然保护区东北马鹿种群遗传多样性与性别结构分析[J]. 兽类学报, 2021, 41(1): 42-50. |
| [10] | 高惠 乔付杰 滕丽微 李俊乐 余梦琦 刘振生. 马鹿阿拉善亚种遗传多样性和群体结构[J]. 兽类学报, 2020, 40(5): 458-466. |
| [11] | 吴采雯 江慧萍 张迪 朱林 郝秀青 张宏科 周煜 乔莹 张学雷 孙璐 杨光 陈炳耀. 北部湾涠洲岛水域发现上颌和须板异常的布氏鲸:对保护的启示[J]. 兽类学报, 2020, 40(4): 329-336. |
| [12] | 刘铸 蒋纹静 王奥男 杨茜 金志民 田新民 李殿伟. 东北地区中鼩鼱遗传多样性与分子系统地理学[J]. 兽类学报, 2020, 40(3): 249-260. |
| [13] | 朱林 李婷 陈炳耀 杨光. 利用形态学及mtDNA基因分子鉴定长须鲸和大村鲸[J]. 兽类学报, 2020, 40(1): 37-46. |
| [14] | 王豆 许冠 王洪永 何森 卜书海 郑雪莉. 中国圈养林麝微卫星DNA多样性研究[J]. 兽类学报, 2019, 39(6): 599-607. |
| [15] | 张于光 Charlotte Hacker 张宇 薛亚东 乌力吉 代云川 罗平 谢然尼玛 Jan E Janecka 李迪强. 三江源和祁连山国家公园雪豹种群的遗传结构分析[J]. 兽类学报, 2019, 39(4): 442-449. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2