

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2025, Vol. 45 ›› Issue (3): 345-355.DOI: 10.16829/j.slxb.150924
唐小澄1,2, 杨婧2, 董艳民3, 张洪亮3, 沈延京3, 王小萌4, 鲍伟东1,2( )
)
收稿日期:2024-03-05
接受日期:2024-09-13
出版日期:2025-05-31
发布日期:2025-06-03
通讯作者:
鲍伟东
作者简介:唐小澄 (1997- ),女,硕士研究生,主要从事动物行为学研究.
基金资助:
Xiaocheng TANG1,2, Jing YANG2, Yanmin DONG3, Hongliang ZHANG3, Yanjing SHEN3, Xiaomeng WANG4, Weidong BAO1,2( )
)
Received:2024-03-05
Accepted:2024-09-13
Online:2025-05-31
Published:2025-06-03
Contact:
Weidong BAO
摘要:
随着北京市自然生态环境逐渐改善,野猪 (Sus scrofa) 的种群数量不断增长,造成局部危害,对野猪种群遗传结构进行监测有助于制定种群长期防控管理措施。本研究使用非损伤采样和微卫星分子标记技术,对北京松山国家级自然保护区野猪种群的性别结构、遗传多样性和遗传距离进行分析,了解种群未来发展趋势。结果表明,88份疑似野猪粪便样品鉴定出42只个体,其中35只雌性、7只雄性,雌雄性比为5∶1。种群遗传多态信息含量 (PIC) 在0.55 ~ 0.81之间,平均为 0.71,具有适中的遗传多样性。基于微卫星位点间的遗传距离构建进化树,42只个体呈现3个分支,聚类分析结果也显示出该种群个体按照3个遗传支系进行聚类的现象。总体上,松山地区的野猪群体在遗传结构方面存在一定程度的分离,但并未到达明显分化的程度,仍属于一个种群。对近交系数的分析显示,松山野猪种群总体呈现中度近交状态,将对种群发展产生一定抑制作用。本研究为了解北京地区野猪种群的遗传结构提供了基础资料,对于分析该地区野猪的种群密度变化和预测种群发展趋势有重要参考价值。
中图分类号:
唐小澄, 杨婧, 董艳民, 张洪亮, 沈延京, 王小萌, 鲍伟东. 北京松山国家级自然保护区野猪种群遗传结构初步分析[J]. 兽类学报, 2025, 45(3): 345-355.
Xiaocheng TANG, Jing YANG, Yanmin DONG, Hongliang ZHANG, Yanjing SHEN, Xiaomeng WANG, Weidong BAO. Genetic structure analysis of wild boar population in Beijing Songshan National Nature Reserve[J]. ACTA THERIOLOGICA SINICA, 2025, 45(3): 345-355.
引物名称 Name | 序列 (5′-3′) Sequence (5′-3′) | 重复单元 Repeating unit/bp | 目的条带大小 Destination stripe size/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| S0090 | CCAAGACTGCCTTGTAGGTGAATA | 2 | 228 ~ 248 | 55 |
| GCTATCAAGTATTGTACCATTAGG | ||||
| SW857 | AGAAATTAGTGCCTCAAATTGG | 2 | 139 ~ 171 | 56 |
| AAACCATTAAGTCCCTAGCAAA | ||||
| S0005 | TCCTTCCCTCCTGGTAACTA | 2 | 202 ~ 244 | 52 |
| GCACTTCCTGATTCTGGGTA | ||||
| SW72 | TGAGAGGTCAGTACAGAAGACC | 2 | 94 ~ 122 | 55 |
| GATCCTCCTCCAAATCCCAT | ||||
| S0226 | AACCTTCCCTTCCCAATCAC | 2 | 86 ~ 114 | 56 |
| CACAGACTGCTTTTTACTCC |
表1 北京松山国家级自然保护区野猪粪样个体识别微卫星引物信息
Table 1 Microsatellite primers for individual identification of wild boars in Beijing Songshan National Nature Reserve
引物名称 Name | 序列 (5′-3′) Sequence (5′-3′) | 重复单元 Repeating unit/bp | 目的条带大小 Destination stripe size/bp | 退火温度 Annealing temperature/℃ |
|---|---|---|---|---|
| S0090 | CCAAGACTGCCTTGTAGGTGAATA | 2 | 228 ~ 248 | 55 |
| GCTATCAAGTATTGTACCATTAGG | ||||
| SW857 | AGAAATTAGTGCCTCAAATTGG | 2 | 139 ~ 171 | 56 |
| AAACCATTAAGTCCCTAGCAAA | ||||
| S0005 | TCCTTCCCTCCTGGTAACTA | 2 | 202 ~ 244 | 52 |
| GCACTTCCTGATTCTGGGTA | ||||
| SW72 | TGAGAGGTCAGTACAGAAGACC | 2 | 94 ~ 122 | 55 |
| GATCCTCCTCCAAATCCCAT | ||||
| S0226 | AACCTTCCCTTCCCAATCAC | 2 | 86 ~ 114 | 56 |
| CACAGACTGCTTTTTACTCC |
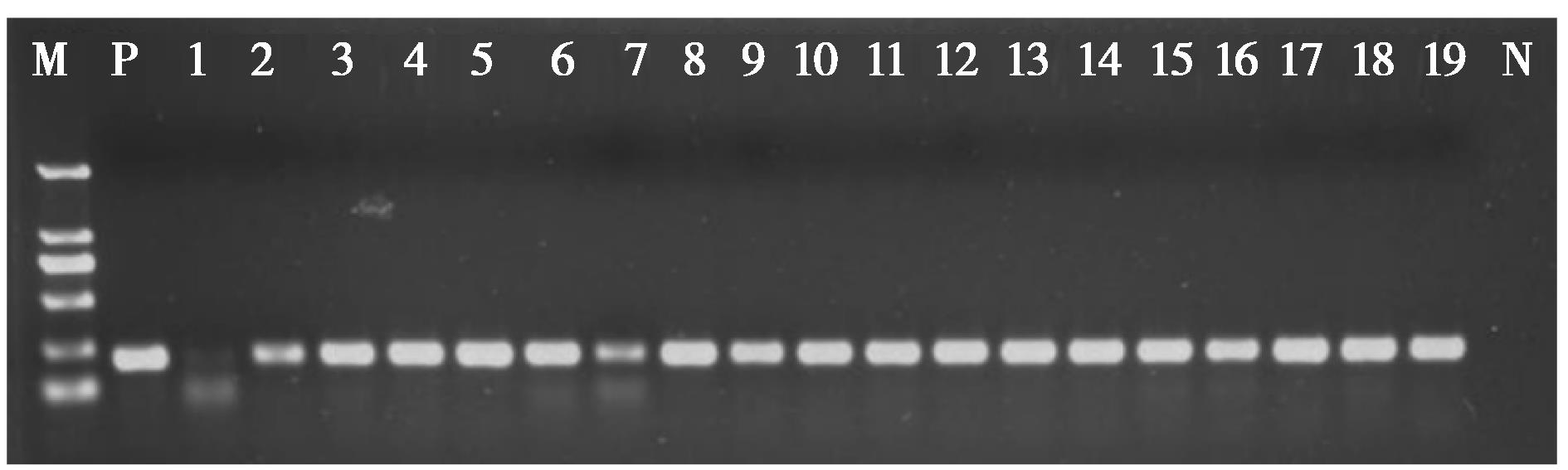
图 2 部分野猪粪样物种识别PCR扩增产物电泳图. M为标记物D2000;P为阳性对照;1 ~ 19为粪便样品DNA;N为阴性对照
Fig. 2 Species identification flectrophoretic photo of PCR amplification products for some of the wild boar fecal samples in Beijing Songshan National Nature Reserve. M represents the marker D2000; P is the positive control; 1 ‑ 19 is the DNA of fecal samples; N is the negative control
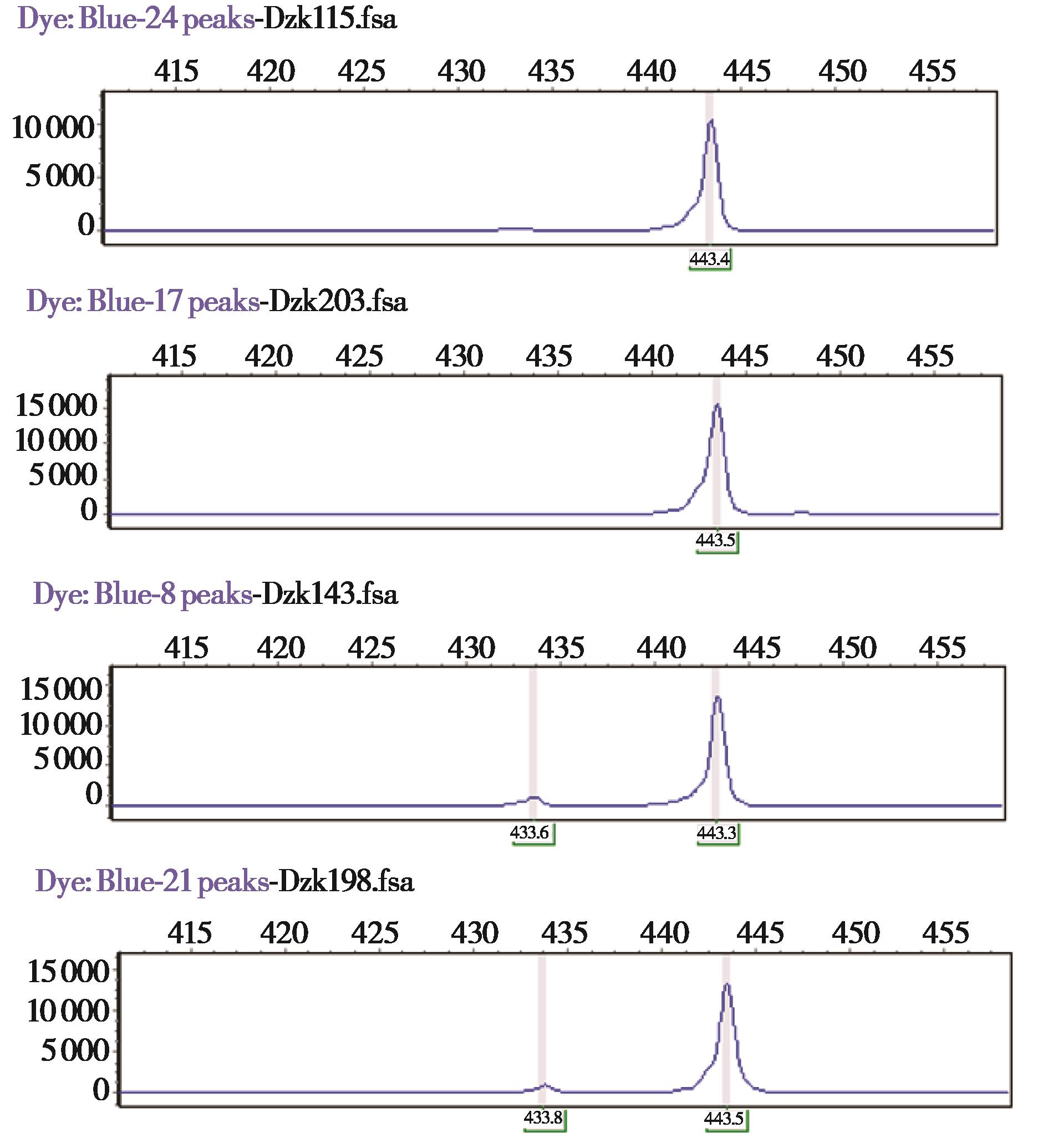
图3 部分野猪性别鉴定PCR扩增产物扫描图. Dzk115和Dzk203为雌性;Dzk143和Dzk198为雄性
Fig. 3 Gene scan of PCR amplification products for sex identification of wild boars in Beijing Songshan National Nature Reserve. Dzk115 and Dzk203 are females; Dzk143 and Dzk198 are males

图4 北京松山国家级自然保护区野猪粪样DNA微卫星座位S0226和SW857的PCR扩增产物扫描图
Fig. 4 Gene scan of PCR amplification products of wild boar microsatellite loci of S0226 and SW857 in Beijing Songshan National Nature Reserve
位点 Locus | 观察等位基因数 Number of alleles (Na ) | 有效等位 基因数 Effective number of allele (Ne ) | 观测 杂合度 Observed heterozygosity (Ho ) | 期望 杂合度 Expected heterozygosity (He ) | 多态信息含量 Polymorphism information content (PIC) |
|---|---|---|---|---|---|
| S0090 | 9 | 4.81 | 0.86 | 0.80 | 0.76 |
| SW857 | 11 | 5.91 | 0.38 | 0.84 | 0.81 |
| S0005 | 9 | 2.44 | 0.60 | 0.60 | 0.55 |
| SW72 | 9 | 3.42 | 0.81 | 0.72 | 0.66 |
| S0226 | 13 | 5.31 | 0.83 | 0.82 | 0.79 |
平均值 Mean | 10.2 | 4.38 | 0.70 | 0.76 | 0.71 |
表2 北京松山国家级自然保护区野猪种群5个微卫星位点的遗传多样性分析
Table 2 Genetic diversity analysis of 5 microsatellite loci in wild boar population in Beijing Songshan National Nature Reserve
位点 Locus | 观察等位基因数 Number of alleles (Na ) | 有效等位 基因数 Effective number of allele (Ne ) | 观测 杂合度 Observed heterozygosity (Ho ) | 期望 杂合度 Expected heterozygosity (He ) | 多态信息含量 Polymorphism information content (PIC) |
|---|---|---|---|---|---|
| S0090 | 9 | 4.81 | 0.86 | 0.80 | 0.76 |
| SW857 | 11 | 5.91 | 0.38 | 0.84 | 0.81 |
| S0005 | 9 | 2.44 | 0.60 | 0.60 | 0.55 |
| SW72 | 9 | 3.42 | 0.81 | 0.72 | 0.66 |
| S0226 | 13 | 5.31 | 0.83 | 0.82 | 0.79 |
平均值 Mean | 10.2 | 4.38 | 0.70 | 0.76 | 0.71 |

图5 基于微卫星共享等位基因距离构建野猪NJ系统发生树. 蓝、黄、红3种颜色区分3个遗传支系;黑色字体代表雌性,紫色字体代表雄性. Dzk:大庄科;Tzg:塘子沟
Fig. 5 The NJ phylogenetic tree of wild boars based on microsatellite shared allele distance. Blue, yellow, and red branches distinguish the three genetic clades; with black fonts representing females and purple fonts representing males. Dzk: Dazhuangke; Tzg: Tangzigou
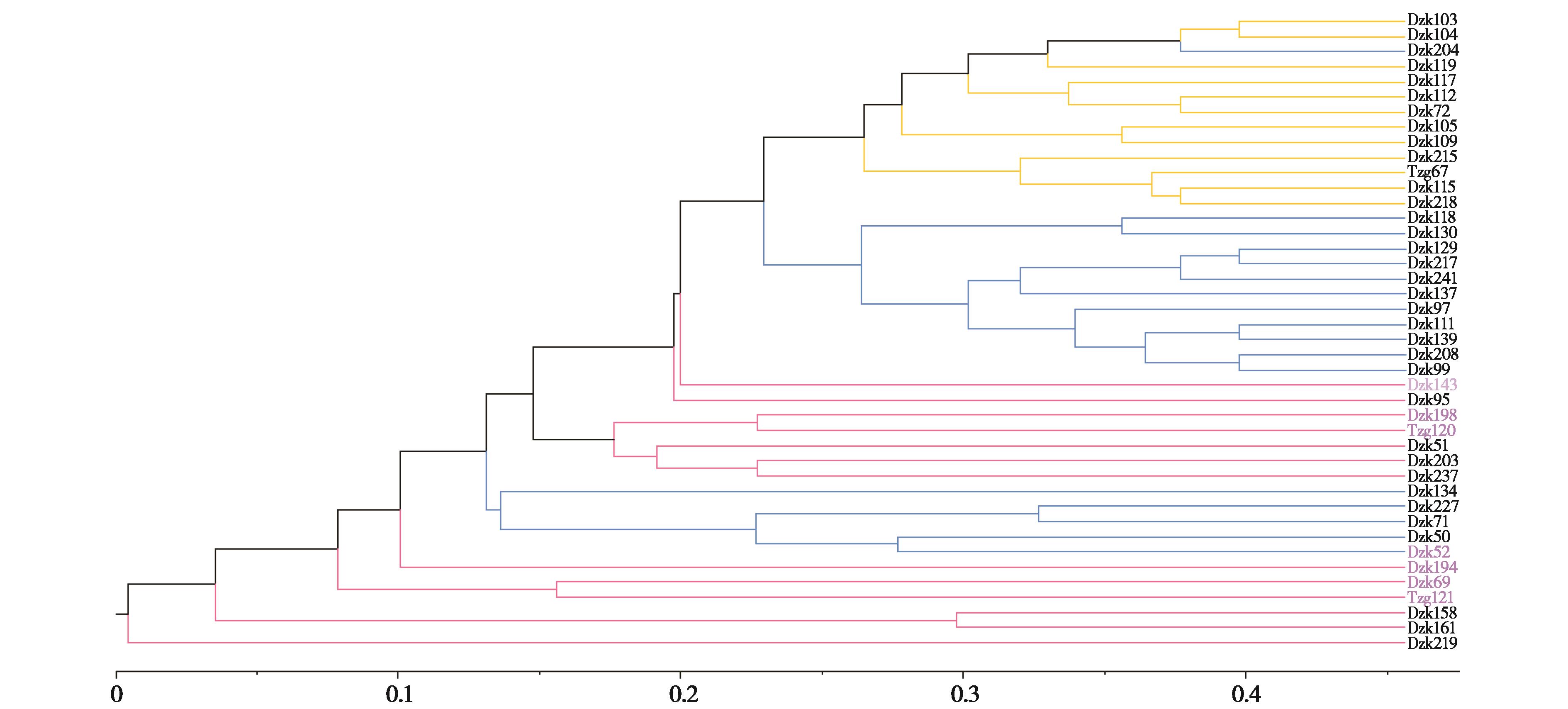
图6 基于微卫星共享等位基因距离构建野猪UPGMA聚类图. 蓝、黄、红3种颜色区分3个遗传支系;黑色字体代表雌性,紫色字体代表雄性. Dzk:大庄科;Tzg:塘子沟
Fig. 6 The clustering tree of wild boars based on microsatellite shared allele distance. Blue, yellow, and red branches distinguish the three genetic clades; with black fonts representing females and purple fonts representing males. Dzk: Dazhuangke; Tzg: Tangzigou
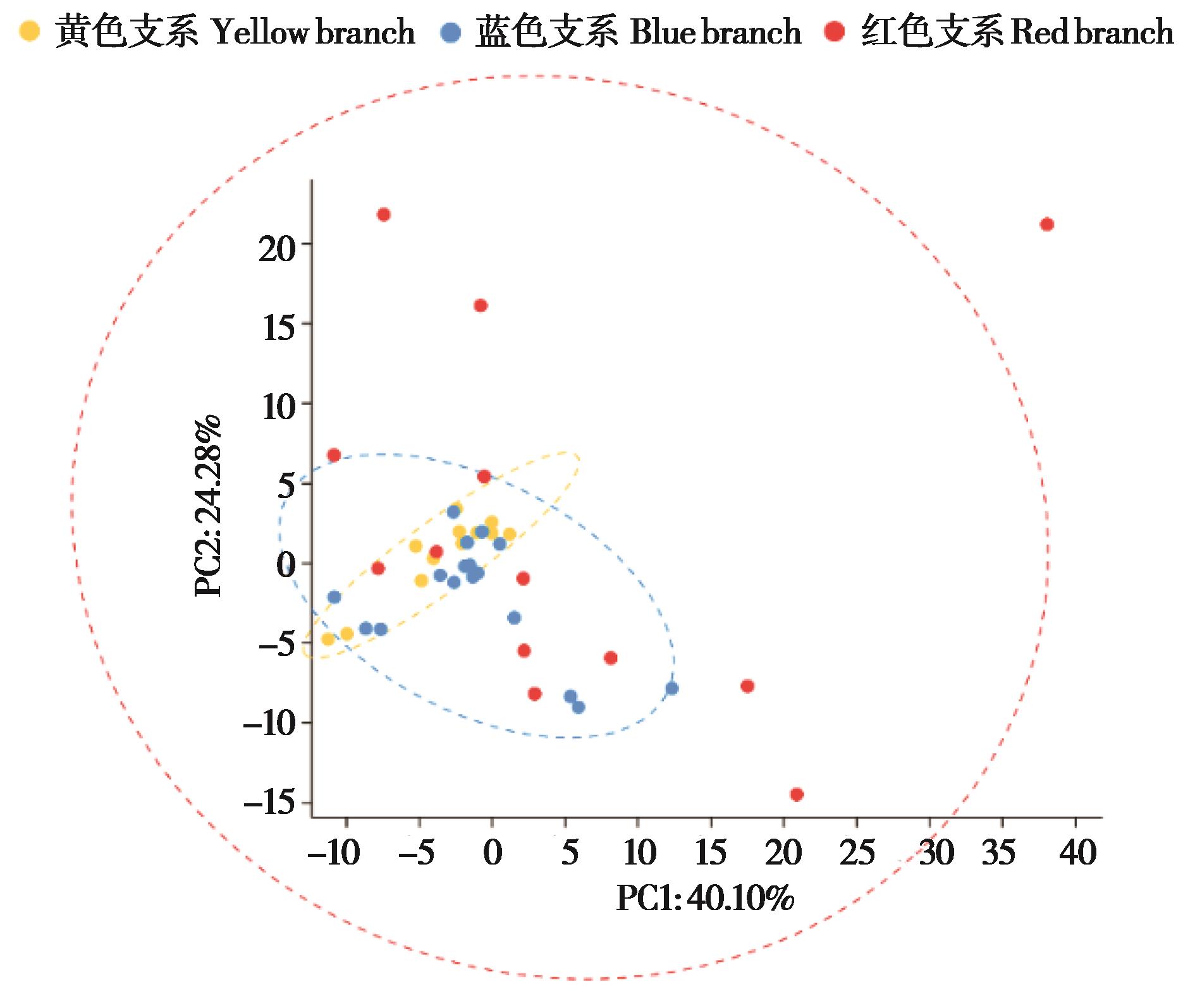
图7 基于微卫星等位基因进行野猪种群的PCA分析. 蓝、黄、红3种颜色区分3个遗传支系
Fig. 7 PCA analysis of wild boar populations based on microsatellite alleles. Blue, yellow, and red branches distinguish the three genetic clades
| Aasen E, Medrano J F. 1990. Amplification of the zfy and zfy genes for sex identification in humans, cattle, sheep and goats [J]. Nature, 8: 1279-1281. | |
| Aziz M A, Tollington S, Barlow A, Greenwood C, Goodrich J M, Smith O, Shamsuddoha M, Islam M A, Groombridge J J. 2017. Using non‑invasively collected genetic data to estimate density and population size of tigers in the Bangladesh Sundarbans [J]. Global Ecology and Conservation, 12: 272-282. | |
| Bellemain E, Swenson J E, Tallmon D, Brunberg S, Taberlet P. 2010. Estimating population size of elusive animals with DNA from hunter‑collected feces: four methods for brown bears [J]. Conservation Biology, 19 (1): 150-161. | |
| Björklund M. 2003. The risk of inbreeding due to habitat loss in the lion (Panthera leo) [J]. Conservation Genetics, 4 (4): 515-523. | |
| Botstein D, White R L, Skolnick M, Davis R W. 1980. Construction of a genetic linkage map in man using restriction fragment length polymorphisms [J]. American Journal of Human Genetics, 32 (3): 314-331. | |
| Du L H, Wang X P, Chen J Q, Liu G L, Wu J G, Zhang Z X. 2012. Comprehensive Scientific Investigation Report on Beijing Songshan Nature Reserve [M]. Beijing: China Forestry Publishing House. (in Chinese) | |
| SoW‑AnGR FAO. 2007. The state of the world’s animal genetic resources for food and agriculture [EB/OL]. Rome: Commission on Genetic Resources for Food and Agriculture, Food and Agriculture Organization of the United Nations. . | |
| Frantz L A F, Schraiber J G, Madsen O, Megens H J, Bosse M, Paudel Y, Semiadi G, Meijaard E, Li Ning, Crooijmans R P M A, Archibald A L, Slatkin M, Schook L B, Larson G, Groenen M A M. 2013. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus [J]. Genome Biology, 14 (9).DOI: 10.1186/gb-2013-14-9-r107 . | |
| Gao J, Guo Z J, Liu Y H. 2016. Biomass allocation pattern and its influencing factors across natural Chinese pine forests of different ages in Songshan, Beijing [J]. Chinese Journal of Ecology, 6: 1475-1480. (in Chinese) | |
| Ge B Q, Hu S X, Sun T Y, Zhang C Y, Jiang G S. 2018. Genetic diversity of wild boar in northern Changbai Mountain [J]. Chinese Journal of Wildlife, 39 (2): 243-249. (in Chinese) | |
| He F, Xu G, Wang X, Liu Y, Liu B Y, Fei Y X. 2023. Habitat selection characteristics of wild boars in Xiaozhaizigou National Nature Reserve, Sichuan Province [J]. Journal of Sichuan Forestry Science and Technology, 44 (2): 13-20. (in Chinese) | |
| He Y J, Yang X L, Ming X Q. 2023. Hazards of living habits and prevention and control countermeasures of wild boar [J]. Agricultural Technology and Equipment, 3: 142-143, 146. (in Chinese) | |
| Hulce D, Li X, Snyderleiby T, Johathan Liu C. 2011. Genemarker® genotyping software: tools to increase the statistical power of DNA fragment analysis [J]. Journal of Biomolecular Techniques, 22 (): 35-36. | |
| Hu Y B, Nie Y G, Wei W, Ma T, Horn R V, Zheng X, Swaisgood R R, Zhou Z, Zhou W, Yan L, Zhang Z J, Wei F W. 2017. Inbreeding and inbreeding avoidance in wild giant pandas [J]. Molecular Ecology, 26 (20): 5793-5806. | |
| Janecka J E, Jackson R, Munkhtsog B, Murphy M J. 2014. Characterization of 9 microsatellites and primers in snow leopards and a species‑specific PCR assay for identifying noninvasive samples [J]. Conservation Genetic Resources, 6: 369-373. | |
| Janecka J E, Tewes M E, Laack L L, Caso A, Grassman LI, Jr Haines A M, Shindle D, Davis B W, Murphy W J, Honeycutt R L. 2015. Reduced genetic diversity and isolation of remnant ocelot populations occupying a severely fragmented landscape in southern Texas [J]. Animal Conservation, 14 (6): 608-619. | |
| Kim B J, Lee H, Lee S. 2010. Species‑ and sex‑specific multiple PCR amplifications of partial cytochrome b gene and Zfx/Zfy introns from invasive and non‑invasive samples of Korean ungulates [J]. Genes and Genomics, 32 (1): 103-104. | |
| Kumar S, Nei M, Dudley J, Tamura K. 2008. Mega: a biologist‑centric software for evolutionary analysis of DNA and protein sequences [J]. Briefings in Bioinformatics, 9 (4): 299-306. | |
| Li C Q, Chang Q, Chen J Q, Zhang B W, Zhu L F, Zhou K Y. 2005. Population structure and phylogeography of the wild boar Sus scrofa in Northeast Asia based on mitochondrial DNA control region variation analysis [J]. Acta Zoologica Sinica, 51 (4):640-649. (in Chinese) | |
| Li H X. 2013. Allelic structure of eight microsatellites of the Amur tiger and their application to population genetic analysis [D]. Ph. D thesis. Harbin: Northeast Forestry University. (in Chinese) | |
| Mayada R F, Tamer S I, Dhama K. 2015. Identification of some domestic animal species (camel, buffalo and sheep) by PCR‑RFLP analysis of the mitochondrial Cytochrome b gene [J]. Advances in Animal and Veterinary Sciences, 3 (2): 136-142. | |
| Miotto R A, Cervini M, Begotti R A, Galetti P M. 2012. Monitoring a puma (Puma concolor) population in a fragmented landscape in Southeast Brazil [J]. Biotropica, 44 (1): 98-104. | |
| Mondol S, Karanth K U, Kumar N S, Gopalaswamy A M, Andheria A, Ramakrishnan U. 2009. Evaluation of non‑invasive genetic sampling methods for estimating tiger population size [J]. Biological Conservation, 142 (10): 2350-2360. | |
| Ning Y. 2020. Study on Amur tiger dispersal, inbreeding and gut microbiota based on DNA analysis [D]. Ph. D thesis. Harbin: Northeast Forestry University. (in Chinese) | |
| Oosterhout C V, Hutchinson W F D, Wills D, Shipley P. 2004. Micro‑checker: Software for identifying and correcting genotyping errors in microsatellite data [J]. Molecular Ecology Notes, 4 (3): 535-538. | |
| Ottoni C, Flink L G, Evin A, Georg C, B de Cupere, van Neer W, Bartosiewicz L, Linderholm A, Barnett R, Peters J, Decorte R, Waelkens M, Vanderheyden N, Ricaut F X, Cakrlar C, Cevik O, Hoelzel A R, Mashkour M, Karimlu A F M, Seno S S. 2013. Pig domestication and human‑mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics [J]. Molecular Biology and Evolution 30 (4): 824-832. | |
| Peakall R, Smouse P E. 2012. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research‑an update [J]. Bioinformatics, 28 (19): 2537-2539. | |
| Peng C C, Li B Q, Wang Y Y, Su H J. 2022. Genetic diversity analysis of Sus scrofa population in Guizhou based on mitochondrial Cyt b gene [J]. China Animal Husbandry and Veterinary Medicine, 49 (7): 2601-2612. (in Chinese) | |
| Pugesek G, Mumma M A, Mahoney S P, Waits L P. 2021. Molecular evaluation of American black bear prey consumption following diversionary feeding [J]. Ursus, 2021 (32e14): 1-11. | |
| Reed D H, Frankham R. 2003. Correlation between fitness and genetic diversity [J]. Conservation Biology, 17 (1): 230-237. | |
| Riordan P. 1998. Unsupervised recognition of individual tigers and snow leopards from their footprints [J]. Animal Forum, 1 (4): 253-262. | |
| Rousset F. 2008. Genepop’007: A complete re‑implementation of the Genepop software for Windows and Linux[CP]. Molecular Ecology Resources, 8 (1): 103-106. | |
| Shen J Y. 2015. Conflict prevention and control technology research of wildlife: a case study in Beijing Songshan National Nature Reserve [D]. Master thesis. Beijing: Beijing Forestry University. (in Chinese) | |
| Spielman D, Brook B W, Frankham R. 2004. Most species are not driven to extinction before genetic factors impact them [J]. Proceedings of the National Academy of Sciences of the United States of America, 101 (42): 15261-15264. | |
| Tian X M, Lian M D, Song Y Q, Liu X H, Yang M P, Chen H. 2022. Genetic diversity and demographic history of Siberian flying squirrel (Pteromys volans) population in northern Zhangguangcai Mountains, Heilongjiang, China [J]. Acta Theriologica Sinica, 42 (4): 398-409. (in Chinese) | |
| Tian X M, Zhang M H. 2023. Genetic diversity of wapiti in northeast China based on fecal DNA [J]. Acta Theriologica Sinica, 43 (1): 41-49. (in Chinese) | |
| Wang C. 2021. Fecal DNA‑based selection of individual recognition microsatellite loci and genetic diversity in Sichuan macaques [D]. Master thesis. Chengdu: Sichuan Agricultural University. (in Chinese) | |
| Wang J L. 2011. COANCESTRY: a program for simulating, estimating and analysing relatedness and inbreeding coefficients [J]. Molecular Ecology Resources, 11: 141-145. | |
| Wang W T. 2020. Analysis of genetic diversity and whole genome sequence in Ussurian wild pig (Sus scrofa ussuricus) [D]. Ph. D thesis. Harbin: Northeast Forestry University. (in Chinese) | |
| Xia F, Bao W D, Gai L X, Ju L F, Huang W H, Jiang J, Ha X B. 2023. Effects of infrastructure and stadium construction on wildlife in Yanqing competition area of Beijing 2022 Olympic Winter Games [J]. Journal of Nanjing Forestry University (Natural Sciences Edition), 47 (1): 221-225. (in Chinese) | |
| Yang G M, Guo Q Y, Yang X W, Peng C C, Zhang M M, Hu C S, Su H J. 2023. Habitat selection of wild boar (Sus scrofa) in mountain area on Guizhou Plateau, China based on camera‑trapping dataset [J]. Acta Ecologica Sinica, 43 (4): 1449-1460. (in Chinese) | |
| Zhang Q, Luo S D Z, Ba S W D, Peng Y Y, E G X, Ni M C R, Suo L D J, Ba D, Dan B, Xian L L, Dan B J S, Zhi Z, Ping C Z D. 2022. Estimation of genetic diversity and population structure of five Tibet yak population using microsatellites markers [J]. China Animal Husbandry and Veterinary Medicine, 49 (6): 2228-2238. (in Chinese) | |
| Zhao G J, Gong Y N, Yang H T, Xie B, Wang T M, Ge J P, Feng L M. 2019. Study on habitat use and activity rhythms of wild boar in eastern region of Northeast Tiger and Leopard National Park [J]. Acta Theriologica Sinica, 39 (4): 431-441. (in Chinese) | |
| Zhao Y J. 2016. Preliminary monitoring of damage caused by wildlife in Beijing Nature Reserves using infrared cameras [D]. Master thesis. Beijing: Beijing Forestry University. (in Chinese) | |
| Zinn H C, Pierce C L. 2002. Values, gender, and concern about potentially dangerous wildlife [J]. Environment and Behavior, 34 (2): 239-256. | |
| 王文涛. 2020. 东北野猪遗传多样性及全基因组序列分析 [D]. 哈尔滨: 东北林业大学博士学位论文. | |
| 王聪. 2021. 基于粪便DNA的川西猕猴个体识别微卫星位点筛选及其遗传多样性研究 [J]. 成都: 四川农业大学硕士学位论文. | |
| 田新民, 张明海. 2023. 基于粪便DNA的东北马鹿遗传多样性 [J]. 兽类学报, 43 (1): 41-49. | |
| 田新民, 廉明栋, 宋雅祺, 刘小慧, 杨孟平, 陈红. 2022. 黑龙江省张广才岭北部小飞鼠的遗传多样性与种群历史动态 [J]. 兽类学报, 42 (4): 398-409. | |
| 宁瑶. 2020. 基于DNA分析的东北虎扩散状况、近交以及肠道菌群的研究 [D]. 哈尔滨: 东北林业大学博士学位论文. | |
| 杜连海, 王小平, 陈峻崎, 刘桂林, 吴记贵, 张志翔. 2012. 北京松山自然保护区综合科学考察报告 [M]. 北京: 中国林业出版社. | |
| 李崇奇, 常青, 陈建琴, 张保卫, 朱立峰, 周开亚. 2005. 东北亚地区野猪种群mtDNA遗传结构及系统地理发生 [J]. 动物学报, 51 (4): 640-649. | |
| 李惠鑫 .2013. 东北虎8个微卫星等位基因结构及群体遗传学分析 [D]. 哈尔滨: 东北林业大学博士学位论文. | |
| 杨光美, 郭群毅, 杨雄威, 彭彩淳, 张明明, 胡灿实, 粟海军. 2023. 基于红外相机数据的贵州高原山地环境野猪生境选择研究 [J]. 生态学报, 43 (4): 1449-1460. | |
| 何仪军, 杨新龙, 明小强. 2023. 野猪的生活习性危害及防控对策 [J]. 农业技术与装备, 3: 142-143, 146. | |
| 沈洁滢. 2015. 野生动物肇事防控技术研究: 以北京松山国家级自然保护区为例 [D]. 北京: 北京林业大学硕士毕业论文. | |
| 张强, 洛桑顿珠, 巴桑旺堆, 彭阳洋, 俄广鑫, 尼玛次仁, 索朗多吉, 巴多, 旦巴, 鲜莉莉, 旦巴加参, 支张, 平措占堆. 2022. 利用微卫星遗传标记评估5个西藏牦牛群体遗传多样性及群体结构 [J]. 中国畜牧兽医, 49 (6): 2228-2238. | |
| 赵永健. 2016. 北京自然保护区肇事野生动物红外相机初步监测[D]. 北京: 北京林业大学硕士学位论文. | |
| 赵国静, 宫一男, 杨海涛, 谢冰, 王天明, 葛剑平, 冯利民. 2019. 东北虎豹国家公园东部的野猪生境利用和活动节律初步研究 [J]. 兽类学报, 39 (4): 431-441. | |
| 贺飞, 许戈, 王鑫, 刘洋, 刘白羽, 费宇翔. 2023. 四川小寨子沟国家级自然保护区野猪生境选择特征 [J]. 四川林业科技, 44 (2): 13-20. | |
| 夏凡, 鲍伟东, 盖立新, 巨龙飞, 黄文华, 蒋健, 哈希博. 2023. 北京冬奥会延庆赛区施工作业对野生动物的影响分析 [J]. 南京林业大学学报 (自然科学版), 47 (1): 221-225. | |
| 高杰, 郭子健, 刘艳红. 2016. 北京松山不同龄级天然油松林生物量分配格局及其影响因子 [J]. 生态学杂志, 6: 1475-1480. | |
| 彭彩淳, 李斌强, 王野影, 粟海军. 2022. 基于线粒体Cyt b基因的贵州野猪 (Sus scrofa) 群体遗传多样性分析 [J].中国畜牧兽医, 49 (7): 2601-2612. | |
| 葛宝庆, 胡素贤, 孙铁铎, 张崇颖, 姜广顺. 2018. 长白山北部林区野猪局域种群的遗传多样性研究 [J]. 野生动物学报, 39 (2): 243-249. |
| [1] | 杜世平, 吴怡豪, 鲁庆斌, 马铭泽, 董爽, 方茹意, 苏秀. 浙江丘陵林业生产区野猪的分布状况及其影响因素[J]. 兽类学报, 2024, 44(4): 466-477. |
| [2] | 陈小南, 田佳, 刘鸣章, 申云逸, 余建平, 刘锋, 申小莉, 李晟. 基于红外相机数据估算浙江省开化县野猪种群数量[J]. 兽类学报, 2023, 43(5): 523-532. |
| [3] | 郑佳鑫, 左清秋, 王刚, 韦旭, 翁晓东, 王正寰. 藏狐的食物组成及其季节差异[J]. 兽类学报, 2023, 43(4): 398-411. |
| [4] | 滕扬, 盖立新, 李建, 房新民, 杨南, 鲍伟东. 两种分析方法在豹猫食物构成中的应用比较[J]. 兽类学报, 2023, 43(1): 50-58. |
| [5] | 饶静, 徐茹洁, 余辉亮, 李巨平, 叶静, 曹胜波, 李翔. 湖北省神农架林区野猪分布与传播非洲猪瘟风险分析[J]. 兽类学报, 2022, 42(2): 189-195. |
| [6] | 谢培根, 胡娟, 李婷婷, 郭瑞, 许丽娟, 宋虓, 李佳琦, 徐爱春. 浙江清凉峰国家级自然保护区野猪空间分布及活动节律[J]. 兽类学报, 2022, 42(2): 168-176. |
| [7] | 杨雄威, 彭彩淳, 郭群毅, 冉景丞, 王野影, 张明明, 胡灿实, 李仕泽, 粟海军. 贵州苗岭地区野猪肠道菌群结构与功能分析[J]. 兽类学报, 2021, 41(4): 365-376. |
| [8] | 王李玲, 胡靖扬, 匡卫民, 于黎. 非损伤性取样样品中富集宿主DNA的研究进展[J]. 兽类学报, 2021, 41(3): 284-295. |
| [9] | 张沼, 张蕊, 李晓宇, 赛罕, 杨振东, 韩志庆, 鲍伟东. 内蒙古赛罕乌拉自然保护区东北马鹿种群遗传多样性与性别结构分析[J]. 兽类学报, 2021, 41(1): 42-50. |
| [10] | 崔爽 刘丙万. 野猪危害防控措施时间延续性及空间推广性研究[J]. 兽类学报, 2020, 40(4): 364-373. |
| [11] | 赵国静 宫一男 杨海涛 谢冰 王天明 葛剑平 冯利民. 东北虎豹国家公园东部的野猪生境利用和活动节律初步研究[J]. 兽类学报, 2019, 39(4): 431-441. |
| [12] | 粟海军 胡灿实 张明明 梁盛. 贵州赤水桫椤国家级自然保护区野猪危害特征与居民态度分析[J]. 兽类学报, 2018, 38(4): 359-368. |
| [13] | 单磊 胡义波 魏辅文. 粪便DNA分析技术在分子生态学研究中的机遇与挑战[J]. 兽类学报, 2018, 38(3): 235-246. |
| [14] | 谢跃 周璇 孙韵 古小彬 杨光友. 基于线粒体12S对中国两大山系大熊猫西氏贝蛔虫种群遗传多样性的分析[J]. 兽类学报, 2015, 35(3): 328-335. |
| [15] | 王长平 刘雪华 武鹏峰 蔡琼 邵小明 朱云 Melissa Songer. 应用红外相机技术研究秦岭观音山自然保护区内野猪的行为和丰富度[J]. 兽类学报, 2015, 35(2): 147-156. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2