

中国核心期刊要目总览
中国科技核心期刊
CSCD数据库源刊
中国科技核心期刊
CSCD数据库源刊


兽类学报 ›› 2022, Vol. 42 ›› Issue (5): 519-530.DOI: 10.16829/j.slxb.150721
张良志1,2, 唐显江1,3, 任世恩1,3, 赵雅琪1,3, 张堰铭1,2( )
)
收稿日期:2022-08-02
接受日期:2022-08-22
出版日期:2022-09-30
发布日期:2022-09-21
通讯作者:
张堰铭
作者简介:张良志 (1981- ),女,副研究员,主要从事分子生态学研究.
基金资助:
Liangzhi ZHANG1,2, Xianjiang TANG1,3, Shien Ren1,3, Yaqi ZHAO1,3, Yanming ZHANG1,2( )
)
Received:2022-08-02
Accepted:2022-08-22
Online:2022-09-30
Published:2022-09-21
Contact:
Yanming ZHANG
摘要:
群落生态构建过程是近年来微生物群落生态学的研究热点。室内饲养可引起肠道菌群的剧烈变化,这种改变是否会影响群落的构建过程,一直未见报道。本文以高原鼠兔 (Ochotona curzoniae) 为对象,采用16S rRNA测序技术,探讨室内饲养和野生高原鼠兔肠道微生物群落在结构、功能以及群落构建过程等方面的差异。结果表明,在繁殖季节,室内饲养组的群落多样性指数和均匀度指数均显著低于野外组;群落丰度指数和群落覆盖度指数在非繁殖季节显著高于繁殖季节。拟杆菌门 (Bacteroidetes) 在室内饲养组显著富集,而厚壁菌门 (Firmicute) 和浮霉菌门 (Planctomycetes) 在野外组显著富集;在野外组,Epsilonbacteraeota和软壁菌门 (Tenericutes) 在繁殖季节显著富集。菌群功能分析显示,室内饲养组与野生组在细胞通讯和心血管疾病通路存在显著差异;野外组繁殖季节与非繁殖季节肠道菌群功能在氨基酸代谢、碳水化合物代谢和脂质代谢等通路存在显著差异。中性模型拟合结果表明,室内饲养明显降低了菌群构建的随机过程。野外组生理状态也会降低菌群构建的随机性。本研究证明室内饲养和宿主生理状态均会对高原鼠兔肠道菌群组成及群落构建产生影响,此结果为实现高原鼠兔室内繁殖提供了新的理论基础。
中图分类号:
张良志, 唐显江, 任世恩, 赵雅琪, 张堰铭. 室内饲养对高原鼠兔肠道微生物群落构建过程的影响[J]. 兽类学报, 2022, 42(5): 519-530.
Liangzhi ZHANG, Xianjiang TANG, Shien Ren, Yaqi ZHAO, Yanming ZHANG. Effects of captivity on the assembly process of microbiota communities of plateau pikas[J]. ACTA THERIOLOGICA SINICA, 2022, 42(5): 519-530.

图1 不同组间高原鼠兔肠道微生物alpha多样性指数差异. a:Sobs指数;b:Ace指数;c:Chao指数;d:Coverage 指数;e:Shannon指数;f: Shannoneven指数
Fig. 1 Differences in alpha diversity of the gut microbiota among three plateau pika groups. a: Sobs index; b: Ace index; c: Chao index; d: Coverage index; e: Shannon index; f: Shannoneven index
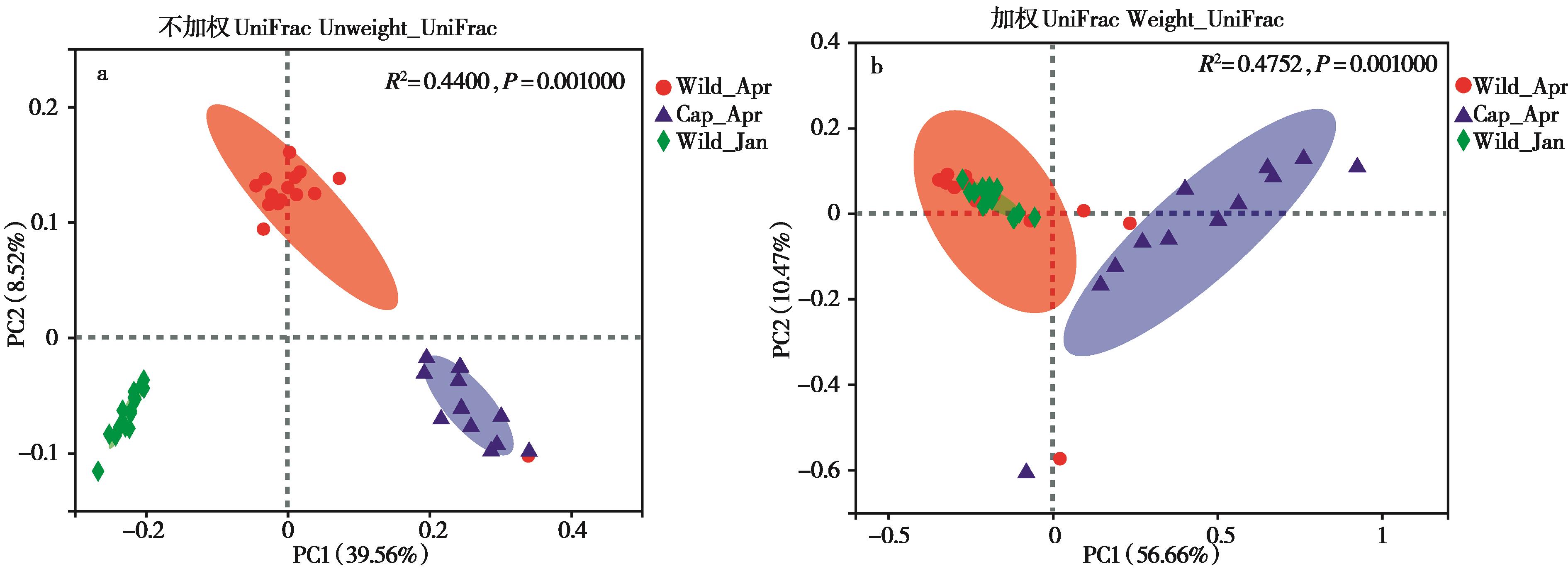
图2 不同组间高原鼠兔肠道微生物结构变化. a:依据不加权UniFrac 距离的PCoA;b:依据加权UniFrac 距离的PCoA
Fig. 2 The structure of the gut microbiota among different plateau pika groups. a: PCoA based on unweighted UniFrac distances; b: PCoA based on weighted UniFrac distances
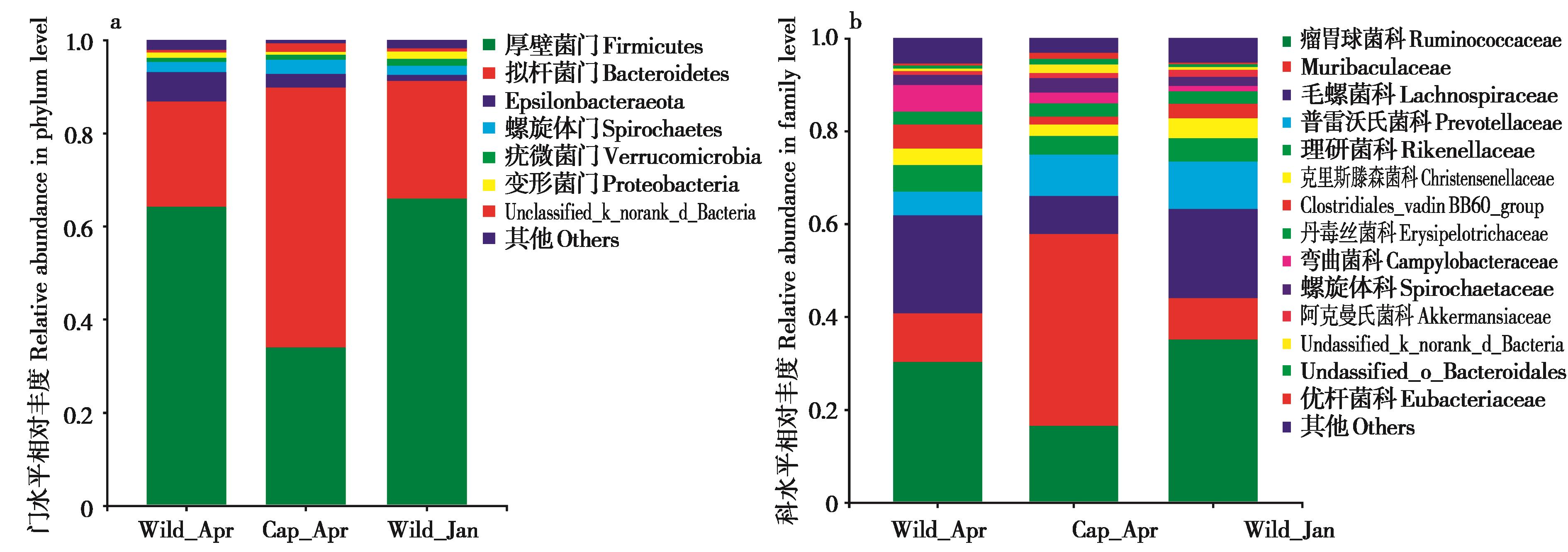
图3 不同组间高原鼠兔肠道微生物组成分析. a:门水平组成;b:科水平组成.
Fig. 3 Differences in taxonomic compositions of the gut microbiota among different plateau pika groups. a: phylum level; b: family level
| OTU ID | Cap_Apr | Wild_Apr | Wild_Jan |
|---|---|---|---|
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes | 0.339 225 | 0.641 988 | 0.659 580 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes | 0.558 989 | 0.225 796 | 0.252 759 |
| d__Bacteria; k__norank_d__Bacteria; p__Epsilonbacteraeota | 0.028 674 | 0.063 268 | 0.012 728 |
| d__Bacteria; k__norank_d__Bacteria; p__Spirochaetes | 0.031 141 | 0.021 820 | 0.019 720 |
| d__Bacteria; k__norank_d__Bacteria; p__Verrucomicrobia | 0.010 711 | 0.009 175 | 0.015 093 |
| d__Bacteria; k__norank_d__Bacteria; p__Proteobacteria | 0.006 001 | 0.011 293 | 0.015 427 |
| d__Bacteria; k__norank_d__Bacteria; p__unclassified_k__norank_d__Bacteria | 0.018 309 | 0.005 707 | 0.006 724 |
| Others | 0.006 950 | 0.020 953 | 0.017 969 |
表1 高原鼠兔肠道菌群门水平相对丰度
Table 1 The percentage of the bacteria in phylum level of plateau pika
| OTU ID | Cap_Apr | Wild_Apr | Wild_Jan |
|---|---|---|---|
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes | 0.339 225 | 0.641 988 | 0.659 580 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes | 0.558 989 | 0.225 796 | 0.252 759 |
| d__Bacteria; k__norank_d__Bacteria; p__Epsilonbacteraeota | 0.028 674 | 0.063 268 | 0.012 728 |
| d__Bacteria; k__norank_d__Bacteria; p__Spirochaetes | 0.031 141 | 0.021 820 | 0.019 720 |
| d__Bacteria; k__norank_d__Bacteria; p__Verrucomicrobia | 0.010 711 | 0.009 175 | 0.015 093 |
| d__Bacteria; k__norank_d__Bacteria; p__Proteobacteria | 0.006 001 | 0.011 293 | 0.015 427 |
| d__Bacteria; k__norank_d__Bacteria; p__unclassified_k__norank_d__Bacteria | 0.018 309 | 0.005 707 | 0.006 724 |
| Others | 0.006 950 | 0.020 953 | 0.017 969 |
| OTU ID | Cap_Apr | Wild_Apr | Wild_Jan |
|---|---|---|---|
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae | 0.165 574 | 0.302 052 | 0.350 869 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales;f__Muribaculaceae | 0.412 680 | 0.104 957 | 0.088 984 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae | 0.081 744 | 0.211 662 | 0.192 503 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales;f__Prevotellaceae | 0.089 237 | 0.050 616 | 0.102 269 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae | 0.040 646 | 0.057 848 | 0.050 388 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Christensenellaceae | 0.023 953 | 0.034 994 | 0.042 735 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiales_vadinBB60_group | 0.017 045 | 0.051 833 | 0.031 483 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae | 0.028 851 | 0.027 822 | 0.026 446 |
d__Bacteria; k__norank_d__Bacteria; p__Epsilonbacteraeota; c__Campylobacteria; o__Campylobacterales; f__Campylobacteraceae | 0.023 531 | 0.057 268 | 0.011 288 |
| d__Bacteria; k__norank_d__Bacteria; p__Spirochaetes; c__Spirochaetia; o__Spirochaetales;f__Spirochaetaceae | 0.031 141 | 0.021 820 | 0.019 720 |
| Others | 0.030 931 | 0.054 559 | 0.052 066 |
表2 高原鼠兔肠道菌群科水平相对丰度
Table 2 The percentage of the bacteria in family level of plateau pika
| OTU ID | Cap_Apr | Wild_Apr | Wild_Jan |
|---|---|---|---|
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Ruminococcaceae | 0.165 574 | 0.302 052 | 0.350 869 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales;f__Muribaculaceae | 0.412 680 | 0.104 957 | 0.088 984 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Lachnospiraceae | 0.081 744 | 0.211 662 | 0.192 503 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales;f__Prevotellaceae | 0.089 237 | 0.050 616 | 0.102 269 |
| d__Bacteria; k__norank_d__Bacteria; p__Bacteroidetes; c__Bacteroidia; o__Bacteroidales; f__Rikenellaceae | 0.040 646 | 0.057 848 | 0.050 388 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Christensenellaceae | 0.023 953 | 0.034 994 | 0.042 735 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Clostridia; o__Clostridiales; f__Clostridiales_vadinBB60_group | 0.017 045 | 0.051 833 | 0.031 483 |
| d__Bacteria; k__norank_d__Bacteria; p__Firmicutes; c__Erysipelotrichia; o__Erysipelotrichales; f__Erysipelotrichaceae | 0.028 851 | 0.027 822 | 0.026 446 |
d__Bacteria; k__norank_d__Bacteria; p__Epsilonbacteraeota; c__Campylobacteria; o__Campylobacterales; f__Campylobacteraceae | 0.023 531 | 0.057 268 | 0.011 288 |
| d__Bacteria; k__norank_d__Bacteria; p__Spirochaetes; c__Spirochaetia; o__Spirochaetales;f__Spirochaetaceae | 0.031 141 | 0.021 820 | 0.019 720 |
| Others | 0.030 931 | 0.054 559 | 0.052 066 |
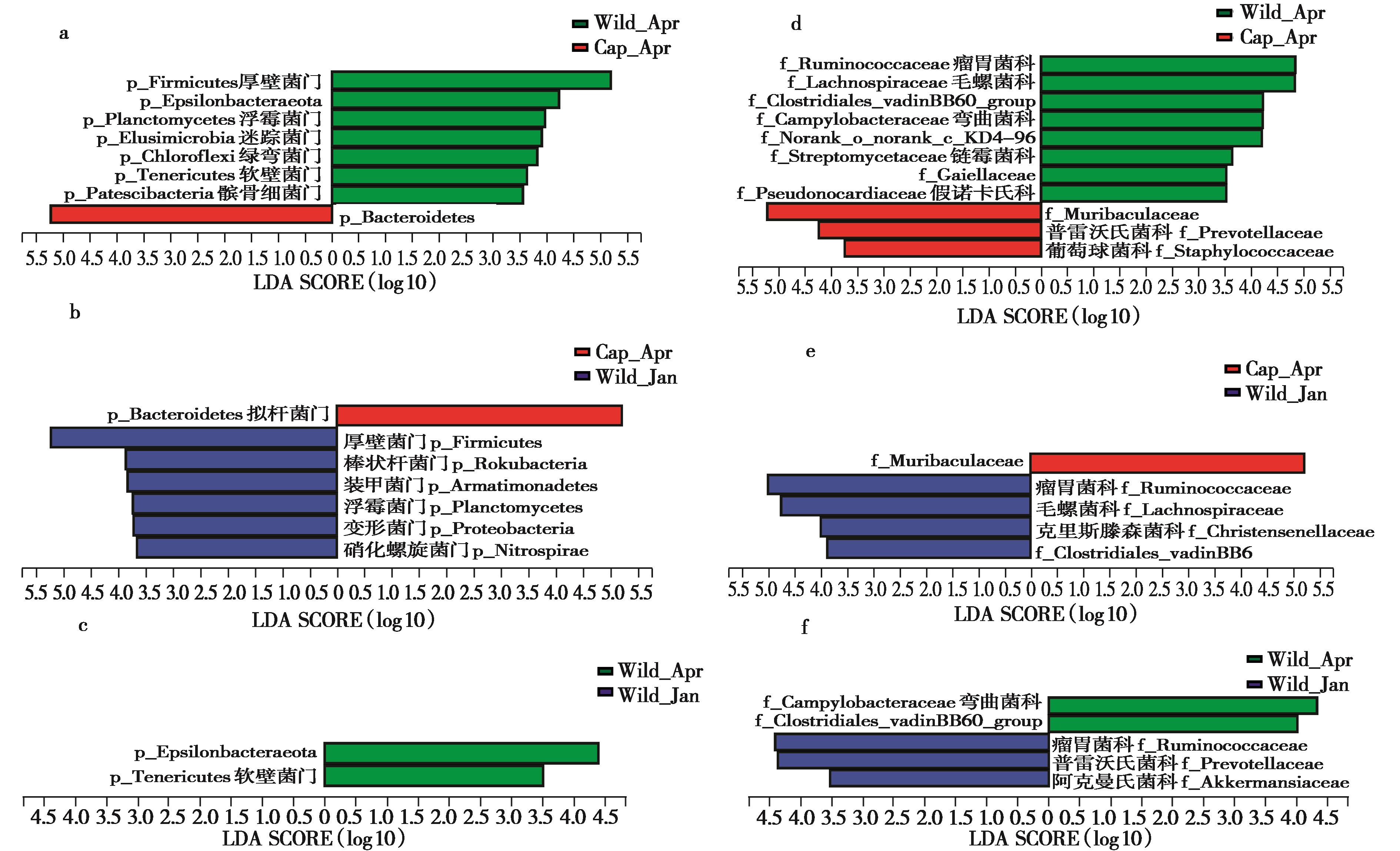
图4 不同组间高原鼠兔肠道微生物相对丰度的差异. a, b, c:门水平的组间两两比较;d, e, f:科水平组间两两比较 (LEfSe, LDA > 4, P < 0.05)
Fig. 4 Taxonomic alterations of the gut microbiota in different plateau pika groups. Pairwise comparisons of Cap_Apr vs. Wild_Apr; Cap_Apr vs. Wild_Jan; Wild_Apr vs. Wild_Jan in phylum level ( a, b, c) and in family level (d, e, f) by the linear discriminant analysis effect size (LEfSe) method (LDA > 4, P < 0.05)
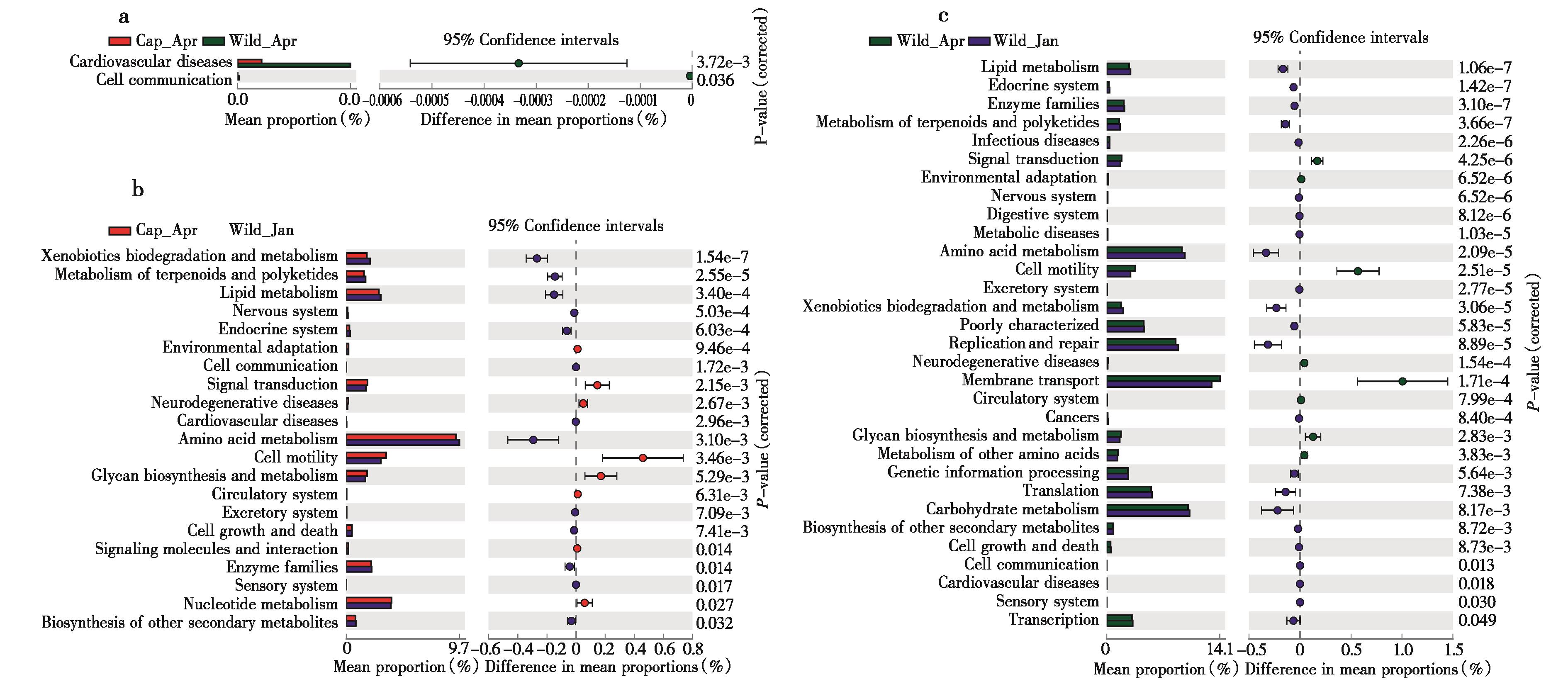
图5 不同组间高原鼠兔肠道微生物菌群功能差异分析. Level 3 水平的KEEG分析,a:Cap_Apr和 Wild_Apr 比较结果;b:Cap_Apr和Wild_Jan 比较结果;c:Wild_Apr 和Wild_Jan比较结果, 使用welch t-test检验
Fig. 5 Functional variations of the gut microbiota in different plateau pika groups. Level 3 KEGG pathways were differentially represented between the Cap_Apr vs. Wild_Apr (a), Cap_Apr vs. Wild_Jan (b), Wild_Apr vs. Wild_Jan (c). Differences were assessed by the Welch t-test and are denoted as the corrected P-value
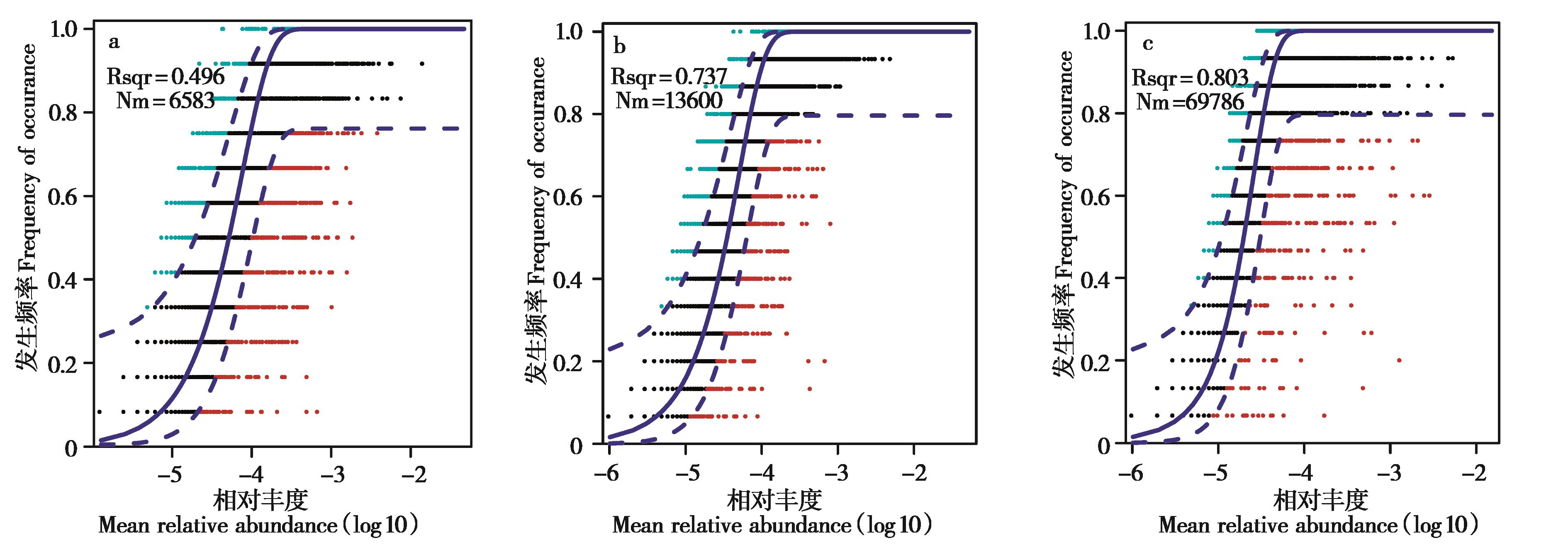
图6 不同组间高原鼠兔肠道微生物菌群构建过程分析. a:Cap_Apr;b:Wild_Apr;c:Wild_Jan
Fig. 6 Assembly process of microbiota communities in different groups of plateau pikas. Predicted occurrence frequencies for Cap_Apr (a), Wild_Apr (b) and Wild_Jan (c)
| 成分 Content | 比例 Percentage(%) |
|---|---|
| 水分 Water | 7.8 |
| 粗蛋白 Crude protein | 16.6 |
| 粗脂肪 Crude fat | 3.1 |
| 粗纤维 Crude fiber | 12.2 |
| 粗灰分 Crude ash | 8.5 |
| 无氮浸出物 Nnitrogen-free extract | 52.8 |
表3 饲养高原鼠兔饲料成分表
Table 3 The content of the artificial diets of captive plateau pikas
| 成分 Content | 比例 Percentage(%) |
|---|---|
| 水分 Water | 7.8 |
| 粗蛋白 Crude protein | 16.6 |
| 粗脂肪 Crude fat | 3.1 |
| 粗纤维 Crude fiber | 12.2 |
| 粗灰分 Crude ash | 8.5 |
| 无氮浸出物 Nnitrogen-free extract | 52.8 |
| Bäckhed F, Ley R E, Sonnenburg J L, Peterson D A, Gordon J I. 2005. Host-bacterial mutualism in the human intestine. Science, 307: 1915-1920. | |
| Bo T B, Zhang X Y, Kohl K D, Wen J, Tian S J, Wang D H. 2020. Coprophagy prevention alters microbiome, metabolism, neurochemistry, and cognitive behavior in a small mammal. ISME Journal, 14 (10): 2625-2645. | |
| Chalifoux L V, Bronson R T, Escajadillo A, McKenna S. 1982. An analysis of the association of gastroenteric lesions with chronic wasting syndrome of marmosets. Veterinary Pathology Supplement, 7: 141-162. | |
| Chase J M. 2010. Stochastic community assembly causes higher biodiversity in more productive environments. Science, 328: 1388-1391. | |
| Chave J. 2004. Neutral theory and community ecology. Ecology Letters, 7: 241-253. | |
| Chesson P. 2000. Mechanisms of maintenance of species diversity. Annual Review Ecology Systematics, 31: 343-366. | |
| Chiyo P I, Grieneisen L E, Wittemyer G, Moss C J, Lee P C, Douglas-Hamilton I, Archie E A. 2014. The influence of social structure, habitat, and host traits on the transmission of Escherichia coli in wild elephants. PLoS ONE, 9 (4): e93408. | |
| Chen S F, Zhou Y Q, Chen Y R, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics, 34: i884-i890. | |
| Clayton J B, Vangay P, Huang H, Ward T, Hillmann B M, Al‑Ghalith G A, Travis D A, Long H T, Tuan B V, Minh V V, Cabana F, Nadler T, Toddes B, Murphy T, Glander K E, Johnson T J, Knights D. 2016. Captivity humanizes the primate microbiome. Proceeding of the National Academy of Sciences of the United States of America, 113: 10376-10381. | |
| Craft M E. 2015. Infectious disease transmission and contact networks in wildlife and livestock. Philosphical Transactions Royal Society B-Biological Science, 370 (1669): 20140107. | |
| Crean A J, Senior A M. 2019. High-fat diets reduce male reproductive success in animal models: a systematic review and meta-analysis. Obesity Reviews, 20: 921-933. | |
| Drake J A. 1990. The mechanics of community assembly and succession. Journal of Theoretical Biology, 147 (2): 213-233. | |
| Edgar R C. 2013. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10: 996-998. | |
| Etienne R S, Olff H. 2004. A novel genealogical approach to neutral biodiversity theory. Ecology Letters, 7: 170-175. | |
| Fan C, Zhang L Z, Jia S G, Tang X J, Fu H B, Li W J, Liu C F, Zhang H, Cheng Q, Zhang Y M. 2022. Seasonal variations in the composition and functional profiles of gut microbiota reflect dietary changes in plateau pikas. Integrative Zoology, 17 (3): 379-395. | |
| Fu H B, Zhang L Z, Chao F, Li W J, Liu C F, Zhang H, Qi C, Zhang Y M. 2021. Sympatric yaks and plateau pikas promote microbial diversity and similarity by the mutual utilization of gut microbiota. Microorganisms, 9 (9): 1890. | |
| Gao H M, Chi X W, Qin W, Wang L, Song P F, Cai Z Y, Zhang J J, Zhang T Z. 2019. Comparison of the gut microbiota composition between the wild and captive Tibetan wild ass (Equus kiang). Journal of Applied Microbiology, 126: 1869-1878. | |
| Guo W, Sudhanshu M, Wang C D, Zhang H M, Ning R H, Kong F L, Zeng B, Zhao J C, Li Y. 2019. Comparative study of gut microbiota in wild and captive giant pandas (Ailuropoda melanoleuca). Genes, 10: 827. | |
| He Z M. 2010. Potentials and prospects: laboratory animal derived from resource animals in China. Chinese Journal of Comparative Medicine, 20: 1-7.(in Chinese) | |
| Hubbell S P. 2001. The Unified Neutral Theory of Biodiversity and Biogeography. Princeton NJ: Princeton University Press. | |
| Hutchinson G E. 1959. Homage to santa rosalia or why are there so many kinds of animals? American Naturalist, 93 (870): 145-159. | |
| Holt C C, van der Giezen M, Daniels C L, Stentiford G D, Bass D. 2020. Spatial and temporal axes impact ecology of the gut microbiome in juvenile European lobster (Homarus gammarus). The ISME Journal, 14: 531-543. | |
| Kau A L, Ahern P P, Griffin N W, Goodman A L, Gordon J I. 2011. Human nutrition, the gut microbiome and the immune system. Nature, 474: 327-336. | |
| Keddy P. 1999. Wetland restoration: The potential for assembly rules in the service of conservation. Wetlands, 19 (4): 716-732. | |
| Khakisahneh S, Zhang X Y, Nouri Z, Wang D H. 2020. Gut microbiota and host thermoregulation in response to ambient temperature fluctuations. Msystems, 5: e00514-e005120. | |
| Kohl K D, Skopec M M, Dearing M D. 2014. Captivity results in disparate loss of gut microbial diversity in closely related hosts. Conservation Physiology, 2 (1): cou009. | |
| Ley R E, Hamady M, Lozupone C A, Turnbaugh P J, Ramey R R, Bircher J S, Schlegel M L, Tucker T A, Schrenzel M D, Knight R, Gordon J I. 2008. Evolution of mammals and their gut microbe. Science, 320: 1647-1651. | |
| Li B, Gao H M, Song P F, Liang C B, Jiang F, Xu B, Liu D X, Zhang T Z. 2022. Captivity shifts gut microbiota communities in white lipped deer (Cervus albirostris). Animals, 12 (4): 431. | |
| Li H, Li T T, Qu J P. 2018. Stochastic processes govern bacterial communities from the blood of pikas and from their arthropod vectors. FEMS Microbiol Ecology, 94 (6): 29722798. | |
| Li Z W, Sun R Y. 1998. Seasonal reproductive cycles in male plateau pika (Ochotona curzoniae). Acta Theriologica Sinica, 18 (1): 42-49 | |
| Liu C B, Hu J Y, Wu Y J, David M I, Chen W, Zhang Z J, Yu L. 2021. Comparative study of gut microbiota from captive and confiscated rescued wild pangolins. Journal of Genetics and Genomics, 48 (9): 825-835. | |
| Langille M G I, Zaneveld J, Caporaso J G, McDonald D, Knights D, Reyes J A, Clemente J C, Burkepile D E, Vega Thurber R L, Knight R, Beiko R G, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nature Biotechnology, 31: 814-821. | |
| MacArthur R H, Wilson E O. 1967. The Theory of Island Biogeography. Princeton NJ: Princeton University Press. | |
| Magoč T, Salzberg S L. 2011. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27: 2957-2963. | |
| Martin L B, Kidd L, Liebl A L, Coon C A. 2011. Captivity induces hyper-inflammation in the house sparrow (Passer domesticus). Journal of Experimental Biology, 214: 2579-2585. | |
| Martínez‑Salinas A, DeClerck F, Vierling K, Vierling L, Legal L, Vílchez‑Mendoza S, Avelino J. 2016. Bird functional diversity supports pest control services in a Costa Rican coffee farm. Agriculture, Ecosystems & Environment, 235: 277-288. | |
| McDonald D, Price M N, Goodrich J, Nawrocki E P, DeSantis T Z, Probst A, Andersen G L, Knight R, Hugenholtz P. 2012. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. The ISME Journal, 6: 610-618. | |
| Miller E A, Johnson T J, Omondi G, Atwill E R, Isbell L A, McCowan B, VanderWaal K. 2018. Assessing transmission of antimicrobial-resistant escherichia coli in wild giraffe contact networks. Applied Environmental Microbiology, 85 (1): e02136-18. | |
| Moeller A H, Suzuki T A, Lin D, Lacey E A, Wasser S K, Nachman M W. 2017. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proceeding of the National Academy of Sciences of the United States of America, 114 (52): 13768-13773. | |
| Neuman H, Debelius J W, Knight R, Koren O. 2015. Microbial endocrinology: the interplay between the microbiota and the endocrine system. FEMS Microbiology Reviews, 39: 509-521. | |
| Ofiţeru I D, Lunn M, Curtis T P, Wells G F, Criddle C S, Francis C A, Sloan W T. 2010. Combined niche and neutral effects in a microbial wastewater treatment community. Proceeding of the National Academy of Sciences of the United States of America, 107: 15345-15350. | |
| Pearson D E, Ortega Y K, Eren Ö, Hierro J L. 2018. Community assembly theory as a framework for biological invasions. Trends in Ecology & Evolution, 33 (5): 313-325. | |
| Pontarp M, Petchey O L. 2016. Community trait overdispersion due to trophic interactions: concerns for assembly process inference. Proceedings of the Royal Society B: Biological Sciences, 283: 20161729. | |
| Peng Q H, Dai Q D, Bai X, Wang Z S, Hu R, Cao G, Li G Y, Wang L Z, Zou H W, Ma J, Zhu Y X. 2021. Comparative study of growth performance, nutrient digestibility, and ruminal and fecal bacterial community between yaks and cattle-yaks raised by stall-feeding. AMB Express, 11 (1): 98. | |
| Redford K H, Jensen D B, Breheny J J. 2012. Integrating the captive and wild. Science, 338: 1157-1158. | |
| Shao Y X. 2003. Laboratory Animals. Nanjing: Southeast University Press.(in Chinese) | |
| Sloan W T, Lunn M, Woodcock S, Head I M, Nee S, Curtis T P. 2006. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environmental Microbiology, 8: 732-740. | |
| Smith A T, Badingqiuying, Wilson M C, Hogan B W. 2019. Functional-trait ecology of the plateau pika Ochotona curzoniae in the Qinghai-Tibetan Plateau. Integrative Zoology, 14: 87-103. | |
| Speakman J R, Chi Q S, Oldakowski L, Fu H B, Fletcher Q E, Hambly C, Togo J, Liu X Y, Piertney S B, Wang X H, Zhang L Z, Redman P, Wang L, Tang G B, Li Y G, Cui J G, Thomson P J, Wang Z L, Glover P, Robertson O C, Zhang Y M, Wang D H. 2021. Surviving winter on the Qinghai-Tibetan Plateau: pikas suppress energy demands and exploit yak feces to survive winter. Proceeding of the National Academy of Sciences of the United States of America, 118 (30): e2100707118. | |
| Stegen J C, Fredrickson J K, Wilkins M J, Konopka A E, Nelson W C, Arntzen E V, Chrisler W B, Chu R K, Danczak R E, Fansler S J, Kennedy D W, Resch C T, Tfaily M. 2016. Groundwater-surface water mixing shifts ecological assembly processes and stimulates organic carbon turnover. Nature Communication, 7: 11237. | |
| Tang G S, Liang X X, Yang M Y, Wang T T, Chen J P, Du W G, Li H, Sun B J. 2020. Captivity influences gut microbiota in Crocodile Lizards (Shinisaurus crocodilurus). Frontiers Microbiology, 11: 550. | |
| Tang X J, Zhang L Z, Fan C, Wang L, Fu H B, Ren S E, Shen W J, Jia S G, Wu G F, Zhang Y M. 2021. Dietary fiber influences bacterial community assembly processes in the gut microbiota of Durco × Bamei crossbred pig. Frontiers Microbiology, 12: 688554. | |
| Tett A, Pasolli E, Masetti G, Ercolini D, Segata N. 2021. Prevotella diversity, niches and interactions with the human host. Nature Reviews Microbiology, 19: 585-599. | |
| Turnbaugh P J, Ley R E, Mahowald M A, Magrini V, Mardis E R, Gordon J I. 2006. An obesity‑associated gut microbiome with increased capacity for energy harvest. Nature, 444: 1027-1031. | |
| Tyakht A V, Kostryukova E S, Popenko A S, Belenikin M S, Pavlenko A V, Larin A K, Karpova I Y, Selezneva O V, Semashko T A, Ospanova E A, Babenko V V, Maev I V, Cheremushkin S V, Kucheryavyy Y A, Shcherbakov P L, Grinevich V B, Efimov O I, Sas E I, Abdulkhakov R A, Abdulkhakov S R, Lyalyukova E A, Livzan M A, Vlassov V V, Sagdeev R Z, Tsukanov V V, Osipenko M F, Kozlova I V, Tkachev A V, Sergienko V I, Alexeev D G, Govorun V M. 2013. Human gut microbiota community structures in urban and rural populations in Russia. Nature Communication, 4: 2469. | |
| Volkov I, Banavar J R, Hubbell S P, Maritan A. 2003. Neutral theory and relative species abundance in ecology. Nature, 424: 1035-1037. | |
| Weiher E, Freund D, Bunton T, Stefanski A, Lee T, Bentivenga S. 2011. Advances, challenges and a developing synthesis of ecological community assembly theory. Philosophical Transactions of the Royal Society B-Biological Sciences, 366: 2403-2413. | |
| Wienemann T, Schmitt‑Wagner D, Meuser K, Segelbacher G, Schink B, Brune A, Berthold P.2011. The bacterial microbiota in the ceca of Capercaillie (Tetrao urogallus) differs between wild and captive birds. Systematic Applied Microbiology, 34: 542-551. | |
| Xiao G H, Liu S, Xiao Y H, Zhu Y, Zhao H B, Li A Q, Li Z L, Feng J. 2019. Seasonal changes in gut microbiota diversity and composition in the greater horseshoe bat. Frontiers in Microbiology, 10: 2247. | |
| Xie H C, Luo Q Y, Chen Z, Ma Y G. Blood indices, blood gas, and sex hormones in plateau pika (Ochotona curzoniae) during breeding season. Chinese Journal of Wildlife, 38 (2): 205-214.(in Chinese) | |
| Yachi S, Loreau M.1999. Biodiversity and ecosystem productivity in a fluctuating environment: the insurance hypothesis. Proceeding of the National Academy of Sciences of the United States of America, 96 (4): 1463-1468. | |
| Yan Q Y, Li J J, Yu Y H, Wang J J, He Z L, Nostrand J D V, Kempher M L, Wu L Y, Wang Y P, Liao L J, Li X H, Wu S, Ni J J, Wang C, Zhou J Z. 2016. Environmental filtering decreases with fish development for the assembly of gut microbiota. Environmental Microbiology, 18: 4739-4754. | |
| Yu Q L, Li G L, Li H. 2022. Two community types occur in gut microbiota of large‑sample wild plateau pikas (Ochotona curzoniae). Integrative Zoology, 17: 366-378. | |
| Zheng Q, Lin Y H. 1996. General situation about laboratory of wild animal. Endemic Diseases Bulletin, 11: 83-87.(in Chinese) | |
| Zhou J Z, Ning D L. 2017. Stochastic community assembly: does It matter in microbial ecology? Microbiology and Molecular Biology Reviews, 81 (4): e00002-17. | |
| 贺争鸣. 2010. 我国资源动物的实验动物化潜力与展望. 中国比较医学杂志, 20: 1-7. | |
| 邵义祥. 2003. 医学实验动物学教程. 南京: 东南大学出版社. | |
| 谢惠春, 罗巧玉, 陈志, 马永贵. 2017. 高原鼠兔繁殖季节血液指标、血气及性激素变化. 野生动物学报, 38 (2): 205-214. | |
| 郑 强, 林永红. 1996. 野生动物实验动物化的研究概况. 地方病通报, 11: 83-87. |
| [1] | 黄小龙, 李海波, 张旭, 程绍传, 晏玉莹, 杨伟, 蒙秉顺, 王丞, 杨杰, 冉景丞. 梵净山同域分布黔金丝猴与藏酋猴的肠道微生物结构差异[J]. 兽类学报, 2024, 44(2): 183-194. |
| [2] | 张佳钰, 安志芳, 王志洁, 陈晓琦, 魏登邦. 高海拔环境抑制高原鼢鼠和高原鼠兔的胆汁酸代谢[J]. 兽类学报, 2023, 43(5): 553-567. |
| [3] | 张璐, 吴学琴, 陈慧青, 董昕, 尚国珍, 吴雁, 边疆晖. 食物蛋白质含量对高原鼠兔免疫功能的影响[J]. 兽类学报, 2023, 43(3): 293-303. |
| [4] | 邹书珍, 罗亚, 程鸣, 王凡, 黎大勇, 康迪, 唐贇. 滇金丝猴肠道微生物四环素抗性基因和肠道环境特征[J]. 兽类学报, 2023, 43(3): 304-314. |
| [5] | 江峰, 宋鹏飞, 张婧捷, 高红梅, 汪海静, 蔡振媛, 刘道鑫, 张同作. 不同养殖场林麝肠道微生物组成和功能的差异[J]. 兽类学报, 2023, 43(2): 129-140. |
| [6] | 郭秋艳, 韦晓, 陆媚静, 范鹏来, 周岐海. 非人灵长类肠道微生物研究进展与展望[J]. 兽类学报, 2023, 43(1): 69-81. |
| [7] | 吴学琴, 陈慧青, 张璐, 尚国珍, 吴雁, 边疆晖. 草地退化影响高原鼠兔血清总IgG水平、肠道寄生物感染及肝脏和肾脏指数[J]. 兽类学报, 2022, 42(5): 531-539. |
| [8] | 朱红娟, 李婧, 王苏芹, 唐琦, 蓝贤勇, 曲家鹏. 不同海拔高原鼠兔个性特征与SERT基因多态性关联[J]. 兽类学报, 2022, 42(5): 540-552. |
| [9] | 张湑泽, 付林, 邹小艳, 都玉蓉. 低氧暴露下高原鼠兔肺组织间隙连接蛋白40表达分析[J]. 兽类学报, 2022, 42(5): 572-578. |
| [10] | 范超, 张良志, 付海波, 刘传发, 李文靖, 张贺, 唐显江, 程琪, 沈文娟, 张堰铭. 高原鼠兔肠道微生物中丰富及稀有类群的季节性特征[J]. 兽类学报, 2021, 41(6): 617-630. |
| [11] | 魏辅文, 黄广平, 樊惠中, 胡义波. 中国濒危兽类保护基因组学和宏基因组学研究进展与展望[J]. 兽类学报, 2021, 41(5): 581-590. |
| [12] | 华铣泽 周睿 叶国辉 花蕊 包达尔罕 花立民. 雄性高原鼠兔长鸣时间分配及其影响因素[J]. 兽类学报, 2020, 40(6): 606-614. |
| [13] | 李双 邹小艳 付林 刘忠浩 白祥慧 都玉蓉 郭松长. 高原鼠兔和昆明白小鼠肺组织结构比较[J]. 兽类学报, 2020, 40(2): 162-169. |
| [14] | 谭春桃 余义博 姜占萍 钟亮 张堰铭 曲家鹏. 不同海拔地区高原鼠兔探究性和静止代谢率的差异[J]. 兽类学报, 2020, 40(1): 27-36. |
| [15] | 王玉军 贾功雪 王婷 王秀蓉 刘忠浩 都玉蓉 马建滨. 光周期-褪黑素信号调控高原鼠兔精原细胞分化和季节性精子发生[J]. 兽类学报, 2020, 40(1): 54-63. |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
 青公网安备 63010402000199号 青ICP备05000010号-2
青公网安备 63010402000199号 青ICP备05000010号-2